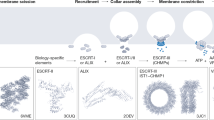
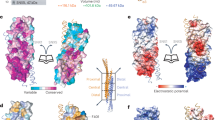
Abstract
The endosomal sorting complex required for transport (ESCRT) machinery is an evolutionarily conserved multisubunit protein complex that remodels cellular membranes. Beyond its classical role in endosomal sorting, the ESCRT machinery has been implicated in an ever-growing number of functions, including viral budding, cytokinesis, autophagy, extracellular vesicle release, pruning of synaptic processes and the repair and closure of holes in cellular membranes. Membrane remodelling functions are typically ascribed to the ESCRT-III subcomplex. In this Review, we discuss recent mechanistic and structural insights into how these proteins assemble and are remodelled to achieve membrane severing. We focus particularly on how ESCRT-III is engaged at different subcellular compartments during both interphase and mitosis to repair and remodel membranes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Katzmann, D. J., Babst, M. & Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 (2001).
Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B. & Emr, S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283–289 (2002).
Babst, M., Katzmann, D. J., Estepa-Sabal, E. J., Meerloo, T. & Emr, S. D. Escrt-III An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271–282 (2002).
Vietri, M., Radulovic, M. & Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020).
Wollert, T. & Hurley, J. H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 (2010).
Raiborg, C. et al. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4, 394–398 (2002).
Mizuno, E., Kawahata, K., Kato, M., Kitamura, N. & Komada, M. STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol. Biol. Cell 14, 3675–3689 (2003).
Luhtala, N. & Odorizzi, G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166, 717–729 (2004).
Agromayor, M. & Martin-Serrano, J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo*. J. Biol. Chem. 281, 23083–23091 (2006).
McCullough, J. et al. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16, 160–165 (2006).
Bowers, K. et al. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII*. J. Biol. Chem. 281, 5094–5105 (2006).
Liu, J. et al. Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. Cell 184, 3660–3673.e18 (2021).
Schlösser, L., Sachse, C., Low, H. H. & Schneider, D. Conserved structures of ESCRT-III superfamily members across domains of life. Trends Biochem. Sci. 48, 993–1004 (2023).
Carlton, J. G. & Baum, B. Roles of ESCRT-III polymers in cell division across the tree of life. Curr. Opin. Cell Biol. 85, 102274 (2023).
Adell, M. A. Y. et al. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 6, e31652 (2017).
Mierzwa, B. E. et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787–798 (2017).
Muzioł, T. et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830 (2006).
Xiao, J. et al. Structural basis of Ist1 function and Ist1–Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20, 3514–3524 (2009).
Bajorek, M. et al. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16, 754–762 (2009).
McCullough, J. et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551 (2015).
Nguyen, H. C. et al. Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat. Struct. Mol. Biol. 27, 392–399 (2020).
Bodon, G. et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J. Biol. Chem. 286, 40276–40286 (2011).
Buchkovich, N. J., Henne, W. M., Tang, S. & Emr, S. D. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell 27, 201–214 (2013).
Huber, S. T., Mostafavi, S., Mortensen, S. A. & Sachse, C. Structure and assembly of ESCRT-III helical Vps24 filaments. Sci. Adv. 6, eaba4897 (2020).
Azad, K. et al. Structural basis of CHMP2A–CHMP3 ESCRT-III polymer assembly and membrane cleavage. Nat. Struct. Mol. Biol. 30, 81–90 (2023).
Webster, B. M. et al. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO 35, 2447–2467 (2016).
Bauer, I., Brune, T., Preiss, R. & Kölling, R. Evidence for a nonendosomal function of the Saccharomyces cerevisiae ESCRT-III-like protein Chm7. Genetics 201, 1439–1452 (2015).
Agromayor, M. et al. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell 20, 1374–1387 (2009).
Clippinger, A. K. et al. IST1 regulates select recycling pathways. Traffic 25, e12921 (2024).
Allison, R. et al. Defects in ER–endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216, 1337–1355 (2017).
Lata, S. et al. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378, 818–827 (2008).
Shim, S., Kimpler, L. A. & Hanson, P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079 (2007).
Strack, B., Calistri, A., Craig, S., Popova, E. & Göttlinger, H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 (2003).
Tang, S. et al. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife 4, e12548 (2015).
Appen, A. et al. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 582, 115–118 (2020).
Tang, S. et al. ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. eLife 5, e15507 (2016).
Gu, M. et al. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl Acad. Sci. USA 114, E2166–E2175 (2017).
Fyfe, I., Schuh, A. L., Edwardson, J. M. & Audhya, A. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J. Biol. Chem. 286, 34262–34270 (2011).
Teis, D., Saksena, S. & Emr, S. D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 (2008).
Teo, H., Perisic, O., González, B. & Williams, R. L. ESCRT-II, an endosome-associated complex required for protein sorting crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 7, 559–569 (2004).
YORIKAWA, C. et al. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 387, 17–26 (2005).
Hanson, P. I., Roth, R., Lin, Y. & Heuser, J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 (2008).
Maity, S. et al. VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci. Adv. 5, eaau7198 (2019).
Glover, J. et al. UMAD1 contributes to ESCRT-III dynamic subunit turnover during cytokinetic abscission. J. Cell Sci. 136, jcs261097 (2023).
Ghazi-Tabatabai, S. et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16, 1345–1356 (2008).
Lata, S. et al. Helical structures of ESCRT-III are disassembled by VPS4. Science 321, 1354–1357 (2008).
Pires, R. et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17, 843–856 (2009).
Effantin, G. et al. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cell. Microbiol. 15, 213–226 (2013).
Shen, Q.-T. et al. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 206, 763–777 (2014).
Chiaruttini, N. et al. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879 (2015).
Henne, W. M., Buchkovich, N. J., Zhao, Y. & Emr, S. D. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151, 356–371 (2012).
Guizetti, J. et al. Cortical constriction during abscission involves helices of ESCRT-III–dependent filaments. Science 331, 1616–1620 (2011).
Goliand, I. et al. Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep. 24, 1756–1764 (2018).
Cashikar, A. G. et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3, e02184 (2014).
Lee, I.-H., Kai, H., Carlson, L.-A., Groves, J. T. & Hurley, J. H. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl Acad. Sci. USA 112, 15892–15897 (2015).
Bertin, A. et al. Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun. 11, 2663 (2020).
Filseck, J. M. et al. Anisotropic ESCRT-III architecture governs helical membrane tube formation. Nat. Commun. 11, 1516 (2020).
Franceschi, N. D. et al. The ESCRT protein CHMP2B acts as a diffusion barrier on reconstituted membrane necks. J. Cell Sci. 132, jcs217968 (2018).
Junglas, B. et al. Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies. Nat. Struct. Mol. Biol. 32, 555–570 (2025).
Naskar, S. et al. Mechanism for Vipp1 spiral formation, ring biogenesis, and membrane repair. Nat. Struct. Mol. Biol. 32, 571–584 (2025).
Pan, S. et al. The cyanobacterial protein VIPP1 forms ESCRT-III-like structures on lipid bilayers. Nat. Struct. Mol. Biol. 32, 543–554 (2025).
Harker-Kirschneck, L., Baum, B. & Šarić, A. Changes in ESCRT-III filament geometry drive membrane remodelling and fission in silico. BMC Biol. 17, 82 (2019).
Jukic, N., Perrino, A. P., Humbert, F., Roux, A. & Scheuring, S. Snf7 spirals sense and alter membrane curvature. Nat. Commun. 13, 2174 (2022).
Moss, F. R. et al. Brominated lipid probes expose structural asymmetries in constricted membranes. Nat. Struct. Mol. Biol. 30, 167–175 (2023).
Scheuring, S. et al. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J. Mol. Biol. 312, 469–480 (2001).
Dvilansky, I., Altaras, Y., Kamenetsky, N., Nachmias, D. & Elia, N. The human AAA-ATPase VPS4A isoform and its co-factor VTA1 have a unique function in regulating mammalian cytokinesis abscission. PLoS Biol. 22, e3002327 (2024).
Das, D. et al. VPS4A is the selective receptor for lipophagy in mice and humans. Mol. Cell 84, 4436–4453.e8 (2024).
Chen, D. et al. VPS4B deficiency causes early embryonic lethality and induces signal transduction disorders of cell endocytosis. Genesis 59, e23415 (2021).
Wenzel, D. M. et al. Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. eLife 11, e77779 (2022).
Monroe, N. et al. The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 426, 510–525 (2014).
Han, H. et al. Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J. Biol. Chem. 290, 13490–13499 (2015).
Monroe, N., Han, H., Shen, P. S., Sundquist, W. I. & Hill, C. P. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife 6, e24487 (2017).
Yang, B., Stjepanovic, G., Shen, Q., Martin, A. & Hurley, J. H. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492–498 (2015).
Scott, A. et al. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl Acad. Sci. USA 102, 13813–13818 (2005).
Stuchell-Brereton, M. D. et al. ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 (2007).
Obita, T. et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 (2007).
Shim, S., Merrill, S. A. & Hanson, P. I. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19, 2661–2672 (2008).
Merrill, S. A. & Hanson, P. I. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition*. J. Biol. Chem. 285, 35428–35438 (2010).
Azmi, I. F. et al. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50–61 (2008).
Vild, C. J., Li, Y., Guo, E. Z., Liu, Y. & Xu, Z. A novel mechanism of regulating the ATPase VPS4 by Its Cofactor LIP5 and the endosomal sorting complex required for transport (ESCRT)-III protein CHMP5*. J. Biol. Chem. 290, 7291–7303 (2015).
Xiao, J. et al. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell 14, 37–49 (2008).
Caillat, C., Maity, S., Miguet, N., Roos, W. H. & Weissenhorn, W. The role of VPS4 in ESCRT-III polymer remodeling. Biochem. Soc. Trans. 47, 441–448 (2019).
Pfitzner, A.-K., Filseck, J. M. von & Roux, A. Principles of membrane remodeling by dynamic ESCRT-III polymers. Trends Cell Biol. 31, 856–868 (2021).
Fabrikant, G. et al. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 5, e1000575 (2009).
Lenz, M., Crow, D. J. G. & Joanny, J.-F. Membrane buckling induced by curved filaments. Phys. Rev. Lett. 103, 038101 (2009).
Liu, M. et al. Three-dimensional architecture of ESCRT-III flat spirals on the membrane. Proc. Natl Acad. Sci. USA 121, e2319115121 (2024).
Schöneberg, J., Lee, I.-H., Iwasa, J. H. & Hurley, J. H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 (2017).
Pfitzner, A.-K. et al. An ESCRT-III polymerization sequence drives membrane deformation and fission. Cell 182, 1140–1155.e18 (2020).
Schöneberg, J. et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362, 1423–1428 (2018).
Ader, N. R. et al. An ESCRT grommet cooperates with a diffusion barrier to maintain nuclear integrity. Nat. Cell Biol. 25, 1465–1477 (2023).
Rue, S. M., Mattei, S., Saksena, S. & Emr, S. D. Novel Ist1-Did2 Complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 (2008).
Dimaano, C., Jones, C. B., Hanono, A., Curtiss, M. & Babst, M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465–474 (2008).
Bajorek, M. et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360–1373 (2009).
Morita, E. et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9, 235–242 (2011).
Kozlovsky, Y. & Kozlov, M. M. Membrane fission: model for intermediate structures. Biophys. J. 85, 85–96 (2003).
Kozlov, M. M., McMahon, H. T. & Chernomordik, L. V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 35, 699–706 (2010).
Johnson, D. S., Bleck, M. & Simon, S. M. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. eLife 7, e36221 (2018).
Cada, A. K. et al. Friction-driven membrane scission by the human ESCRT-III proteins CHMP1B and IST1. Proc. Natl. Acad. Sci. USA 119, e2204536119 (2022).
Flower, T. G. et al. A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission. Nat. Struct. Mol. Biol. 27, 570–580 (2020).
Wang, Y. et al. Biomolecular condensates mediate bending and scission of endosome membranes. Nature 634, 1204–1210 (2024).
King, M. C., Lusk, C. P. & Ader, N. R. Sense, plug, and seal: proteins as both rapid responders and constitutive barriers supporting organelle compartmentalization. Mol. Biol. Cell 36, pe6 (2025).
Denais, C. M. et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016).
Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016).
Hatch, E. M. & Hetzer, M. W. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215, 27–36 (2016).
Zhang, Q. et al. Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Biol. Cell 30, 899–906 (2019).
Kovacs, M. T. et al. DNA damage induces nuclear envelope rupture through ATR-mediated phosphorylation of lamin A/C. Mol. Cell 83, 3659–3668.e10 (2023).
Chen, N. Y. et al. Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl Acad. Sci. USA 115, 10100–10105 (2018).
Heerden, D. V., Klima, S. & Bout, I. V. D. How nuclear envelope dynamics can direct laminopathy phenotypes. Curr. Opin. Cell Biol. 86, 102290 (2024).
Olmos, Y., Hodgson, L., Mantell, J., Verkade, P. & Carlton, J. G. ESCRT-III controls nuclear envelope reformation. Nature 522, 236–239 (2015).
Vietri, M. et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235 (2015).
Halfmann, C. T. et al. Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol. 218, 2136–2149 (2019).
Haraguchi, T. et al. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114, 4575–4585 (2001).
Haraguchi, T. et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121, 2540–2554 (2008).
Samwer, M. et al. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170, 956–972.e23 (2017).
Olmos, Y., Perdrix-Rosell, A. & Carlton, J. G. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 26, 2635–2641 (2016).
Pieper, G. H., Sprenger, S., Teis, D. & Oliferenko, S. ESCRT-III/Vps4 controls heterochromatin-nuclear envelope attachments. Dev. Cell 53, 27–41.e6 (2020).
Barger, S. R., Penfield, L. & Bahmanyar, S. Nuclear envelope assembly relies on CHMP-7 in the absence of BAF–LEM-mediated hole closure. J. Cell Sci. 136, jcs261385 (2023).
Thaller, D. J. et al. Direct binding of ESCRT protein Chm7 to phosphatidic acid-rich membranes at nuclear envelope herniations. J. Cell Biol. 220, e202004222 (2021).
Wallis, S. S. et al. The ESCRT machinery counteracts Nesprin-2G-mediated mechanical forces during nuclear envelope repair. Dev. Cell 56, 3192–3202.e8 (2021).
Thaller, D. J. et al. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife 8, e45284 (2019).
Schwedler et al. The protein network of HIV budding. Cell 114, 701–713 (2003).
Adell, M. A. Y. et al. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 205, 33–49 (2014).
Krupina, K., Goginashvili, A. & Cleveland, D. W. Causes and consequences of micronuclei. Curr. Opin. Cell Biol. 70, 91–99 (2021).
Liu, S. et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561, 551–555 (2018).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Willan, J. et al. ESCRT-III is necessary for the integrity of the nuclear envelope in micronuclei but is aberrant at ruptured micronuclear envelopes generating damage. Oncogenesis 8, 29 (2019).
Vietri, M. et al. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat. Cell Biol. 22, 856–867 (2020).
Gatta, A. T. et al. CDK1 controls CHMP7-dependent nuclear envelope reformation. eLife 10, e59999 (2021).
Bona, M. D. et al. Micronuclear collapse from oxidative damage. Science 385, eadj8691 (2024).
Martin, S. et al. A p62-dependent rheostat dictates micronuclei catastrophe and chromosome rearrangements. Science 385, eadj7446 (2024).
Cheng, X., Zhang, X., Yu, L. & Xu, H. Calcium signaling in membrane repair. Semin. Cell Dev. Biol. 45, 24–31 (2015).
Jimenez, A. J. et al. ESCRT machinery is required for plasma membrane repair. Science 343, 1247136 (2014).
Scheffer, L. L. et al. Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5, 5646 (2014).
Sønder, S. L. et al. Annexin A7 is required for ESCRT III-mediated plasma membrane repair. Sci. Rep. 9, 6726 (2019).
Gong, Y.-N. et al. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 169, 286–300.e16 (2017).
Rühl, S. et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 362, 956–960 (2018).
Ritter, A. T. et al. ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science 376, 377–382 (2022).
Stefani, C. et al. LITAF protects against pore-forming protein-induced cell death by promoting membrane repair. Sci. Immunol. 9, eabq6541 (2024).
Rodríguez-Silvestre, P. et al. Perforin-2 is a pore-forming effector of endocytic escape in cross-presenting dendritic cells. Science 380, 1258–1265 (2023).
Skowyra, M. L., Schlesinger, P. H., Naismith, T. V. & Hanson, P. I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018).
Radulovic, M. et al. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, EMBJ201899753 (2018).
Shukla, S., Larsen, K. P., Ou, C., Rose, K. & Hurley, J. H. In vitro reconstitution of calcium-dependent recruitment of the human ESCRT machinery in lysosomal membrane repair. Proc. Natl Acad. Sci. USA 119, e2205590119 (2022).
Shukla, S. et al. Mechanism and cellular function of direct membrane binding by the ESCRT and ERES-associated Ca2+-sensor ALG-2. Proc. Natl Acad. Sci. USA 121, e2318046121 (2024).
Chen, W., Motsinger, M. M., Li, J., Bohannon, K. P. & Hanson, P. I. Ca2+-sensor ALG-2 engages ESCRTs to enhance lysosomal membrane resilience to osmotic stress. Proc. Natl Acad. Sci. USA 121, e2318412121 (2024).
Mercier, V. et al. Endosomal membrane tension regulates ESCRT-III-dependent intra-lumenal vesicle formation. Nat. Cell Biol. 22, 947–959 (2020).
Rose, K. et al. Tau fibrils induce nanoscale membrane damage and nucleate cytosolic tau at lysosomes. Proc. Natl Acad. Sci. USA 121, e2315690121 (2024).
Chen, J. J. et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 294, 18952–18966 (2019).
Bussi, C. et al. Stress granules plug and stabilize damaged endolysosomal membranes. Nature 623, 1062–1069 (2023).
Ebstrup, M. L. et al. Annexin A7 mediates lysosome repair independently of ESCRT-III. Front. Cell Dev. Biol. 11, 1211498 (2024).
Radulovic, M., Yang, C. & Stenmark, H. Lysosomal membrane homeostasis and its importance in physiology and disease. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-025-00873-w (2025).
Bohannon, K. P. & Hanson, P. I. ESCRT puts its thumb on the nanoscale: fixing tiny holes in endolysosomes. Curr. Opin. Cell Biol. 65, 122–130 (2020).
Nozawa, T. et al. Rab41-mediated ESCRT machinery repairs membrane rupture by a bacterial toxin in xenophagy. Nat. Commun. 14, 6230 (2023).
Yamamoto, H., Zhang, S. & Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 24, 382–400 (2023).
Filimonenko, M. et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 (2007).
Lee, J.-A., Beigneux, A., Ahmad, S. T., Young, S. G. & Gao, F.-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17, 1561–1567 (2007).
Rusten, T. E. et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17, 1817–1825 (2007).
Takahashi, Y. et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9, 2855 (2018).
Zhen, Y. et al. ESCRT-mediated phagophore sealing during mitophagy. Autophagy 16, 826–841 (2020).
Takahashi, Y. et al. VPS37A directs ESCRT recruitment for phagophore closure. J. Cell Biol. 218, 3336–3354 (2019).
Zhou, F. et al. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 218, 1908–1927 (2019).
Liao, Y.-C. et al. COPII with ALG2 and ESCRTs control lysosome-dependent microautophagy of ER exit sites. Dev. Cell 59, 1410–1424.e4 (2024).
Wang, W. et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl Acad. Sci. USA 112, E1373–E1381 (2015).
McGourty, C. A. et al. Regulation of the CUL3 ubiquitin ligase by a calcium-dependent co-adaptor. Cell 167, 525–538.e14 (2016).
Lie-Jensen, A. et al. Centralspindlin recruits ALIX to the midbody during cytokinetic abscission in drosophila via a mechanism analogous to virus budding. Curr. Biol. 29, 3538–3548.e7 (2019).
Carlton, J. G. & Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 (2007).
Morita, E. et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 (2007).
Tedeschi, A. et al. Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11, 1746 (2020).
Goliand, I., Nachmias, D., Ofir, G. & Elia, N. Inhibition of ESCRT II-CHMP6 interactions impede cytokinetic abscission and leads to cell death. Mol. Biol. Cell 25, mbc.E14–08-1317 (2014).
Christ, L. et al. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212, 499–513 (2016).
Elia, N., Sougrat, R., Spurlin, T. A., Hurley, J. H. & Lippincott-Schwartz, J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl Acad. Sci. USA 108, 4846–4851 (2011).
Schiel, J. A. et al. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol. 14, 1068–1078 (2012).
Frémont, S. et al. Oxidation of F-actin controls the terminal steps of cytokinesis. Nat. Commun. 8, 14528 (2017).
Addi, C. et al. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat. Commun. 11, 1941 (2020).
Connell, J. W., Lindon, C., Luzio, J. P. & Reid, E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic 10, 42–56 (2009).
Advedissian, T., Frémont, S. & Echard, A. Cytokinetic abscission requires actin-dependent microtubule severing. Nat. Commun. 15, 1949 (2024).
Yang, D. et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278–1286 (2008).
Eikenes, ÅH. et al. ALIX and ESCRT-III coordinately control cytokinetic abscission during germline stem cell division in vivo. PLoS Genet. 11, e1004904 (2015).
Matias, N. R., Mathieu, J. & Huynh, J.-R. Abscission is regulated by the ESCRT-III protein shrub in drosophila germline stem cells. PLoS Genet. 11, e1004653 (2015).
Hermant, C., Matias, N. R., Michel-Hissier, P., Huynh, J.-R. & Mathieu, J. Lethal giant disc is a target of Cdk1 and regulates ESCRT-III localization during germline stem cell abscission. Development 151, dev202306 (2024).
Carlton, J. G., Caballe, A., Agromayor, M., Kloc, M. & Martin-Serrano, J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336, 220–225 (2012).
Capalbo, L. et al. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open. Biol. 2, 120070 (2012).
Mackay, D. R., Makise, M. & Ullman, K. S. Defects in nuclear pore assembly lead to activation of an Aurora B–mediated abscission checkpoint. J. Cell Biol. 191, 923–931 (2010).
Mackay, D. R. & Ullman, K. S. ATR and a Chk1-Aurora B pathway coordinate postmitotic genome surveillance with cytokinetic abscission. Mol. Biol. Cell 26, 2217–2226 (2015).
Strohacker, L. K. et al. Identification of abscission checkpoint bodies as structures that regulate ESCRT factors to control abscission timing. eLife 10, e63743 (2021).
Thoresen, S. B. et al. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat. Cell Biol. 16, 547–557 (2014).
Lafaurie-Janvore, J. et al. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science 339, 1625–1629 (2013).
Andrade, V. et al. Caveolae promote successful abscission by controlling intercellular bridge tension during cytokinesis. Sci. Adv. 8, eabm5095 (2022).
McMillan, B. J. et al. Structural basis for regulation of ESCRT-III complexes by lgd. Cell Rep. 19, 1750–1757 (2017).
Martinelli, N. et al. CC2D1A is a regulator of ESCRT-III CHMP4B. J. Mol. Biol. 419, 75–88 (2012).
Ventimiglia, L. N. et al. CC2D1B coordinates ESCRT-III activity during the mitotic reformation of the nuclear envelope. Dev. Cell 47, 547–563.e6 (2018).
Raiborg, C. et al. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114, 2255–2263 (2001).
Katzmann, D. J., Stefan, C. J., Babst, M. & Emr, S. D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162, 413–423 (2003).
Merigliano, C. et al. AKTIP interacts with ESCRT I and is needed for the recruitment of ESCRT III subunits to the midbody. PLoS Genet. 17, e1009757 (2021).
Caballe, A. et al. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. eLife 4, e06547 (2015).
Paine, E. L. et al. The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission. eLife 12, e84515 (2023).
Richard, A. et al. Methylation of ESCRT-III components regulates the timing of cytokinetic abscission. Nat. Commun. 15, 4023 (2024).
Crespo-Yàñez, X. et al. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLoS Genet. 14, e1007456 (2018).
Mathieu, J., Michel-Hissier, P., Boucherit, V. & Huynh, J.-R. The deubiquitinase USP8 targets ESCRT-III to promote incomplete cell division. Science 376, 818–823 (2022).
Hurtig, F. et al. The patterned assembly and stepwise Vps4-mediated disassembly of composite ESCRT-III polymers drives archaeal cell division. Sci. Adv. 9, eade5224 (2023).
Pulschen, A. A. et al. Live imaging of a hyperthermophilic archaeon reveals distinct roles for two ESCRT-III homologs in ensuring a robust and symmetric division. Curr. Biol. 30, 2852–2859.e4 (2020).
Risa, G. T. et al. The proteasome controls ESCRT-III–mediated cell division in an archaeon. Science 369, eaaz2532 (2020).
Clayton, E. L. et al. Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 130, 511–523 (2015).
Skibinski, G. et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 (2005).
Holm, I. E., Englund, E., Mackenzie, I. R. A., Johannsen, P. & Isaacs, A. M. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J. Neuropathol. Exp. Neurol. 66, 884–891 (2007).
Deng, X. et al. CHMP2B regulates TDP-43 phosphorylation and cytotoxicity independent of autophagy via CK1. J. Cell Biol. 221, e202103033 (2021).
Belly, A. et al. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J. Cell Sci. 123, 2943–2954 (2010).
Chassefeyre, R. et al. Regulation of postsynaptic function by the dementia-related ESCRT-III subunit CHMP2B. J. Neurosci. 35, 3155–3173 (2015).
Clayton, E. L., Bonnycastle, K., Isaacs, A. M., Cousin, M. A. & Schorge, S. A novel synaptopathy-defective synaptic vesicle protein trafficking in the mutant CHMP2B mouse model of frontotemporal dementia. J. Neurochem. 160, 412–425 (2022).
Coyne, A. N. et al. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 13, eabe1923 (2021).
Rodger, C. et al. De novo VPS4A mutations cause multisystem disease with abnormal neurodevelopment. Am. J. Hum. Genet. 107, 1129–1148 (2020).
Scourfield, E. J. & Martin-Serrano, J. Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans. 45, 613–634 (2017).
Rheinemann, L. et al. RetroCHMP3 blocks budding of enveloped viruses without blocking cytokinesis. Cell 184, 5419–5431.e16 (2021).
Arii, J. et al. ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat. Commun. 9, 3379 (2018).
Mattissek, C. & Teis, D. The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis. Mol. Membr. Biol. 31, 111–119 (2014).
Szymańska, E. et al. Synthetic lethality between VPS4A and VPS4B triggers an inflammatory response in colorectal cancer. EMBO Mol. Med. 12, EMMM201910812 (2020).
Neggers, J. E. et al. Synthetic lethal interaction between the ESCRT paralog enzymes VPS4A and VPS4B in cancers harboring loss of chromosome 18q or 16q. Cell Rep. 33, 108493 (2020).
Kolmus, K. et al. Concurrent depletion of Vps37 proteins evokes ESCRT-I destabilization and profound cellular stress responses. J. Cell Sci. 134, jcs250951 (2021).
Bernareggi, D. et al. CHMP2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity. Nat. Commun. 13, 1899 (2022).
Zheng, Y. et al. CHMP3 promotes the progression of hepatocellular carcinoma by inhibiting caspase-1-dependent pyroptosis. Int. J. Oncol. 64, 8 (2023).
Song, S. et al. CHMP4A stimulates CD8+ T-lymphocyte infiltration and inhibits breast tumor growth via the LSD1/IFNβ axis. Cancer Sci. 114, 3162–3175 (2023).
Lin, S., Wang, M., Cao, Q. & Li, Q. Chromatin modified protein 4C (CHMP4C) facilitates the malignant development of cervical cancer cells. FEBS Open. Bio 10, 1295–1303 (2020).
Yu, L. et al. CHMP4C promotes pancreatic cancer progression by inhibiting necroptosis via the RIPK1/RIPK3/MLKL pathway. J. Adv. Res. https://doi.org/10.1016/j.jare.2025.01.040 (2025).
Sadler, J. B. A. et al. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc. Natl Acad. Sci. USA 115, E8900–E8908 (2018).
Umphred-Wilson, K. et al. The ESCRT protein CHMP5 promotes T cell leukemia by enabling BRD4-p300-dependent transcription. Nat. Commun. 16, 4133 (2025).
Monypenny, J. et al. ALIX regulates tumor-mediated immunosuppression by controlling EGFR activity and PD-L1 presentation. Cell Rep. 24, 630–641 (2018).
Baietti, M. F. et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012).
Yeat, N. Y., Liu, L.-H., Chang, Y.-H., Lai, C. P.-K. & Chen, R.-H. Bro1 proteins determine tumor immune evasion and metastasis by controlling secretion or degradation of multivesicular bodies. Dev. Cell 60, 2114–2130 (2025).
Shiels, A. et al. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am. J. Hum. Genet. 81, 596–606 (2007).
Gulluni, F. et al. PI(3,4)P2-mediated cytokinetic abscission prevents early senescence and cataract formation. Science 374, eabk0410 (2021).
Sagona, A. P., Nezis, I. P. & Stenmark, H. Association of CHMP4B and autophagy with micronuclei: implications for cataract formation. BioMed. Res. Int. 2014, 974393 (2014).
Zhou, Y., Bennett, T. M. & Shiels, A. A charged multivesicular body protein (CHMP4B) is required for lens growth and differentiation. Differentiation 109, 16–27 (2019).
Acknowledgements
J.G.C. is a Wellcome Trust Senior Research Fellow (grant no. 224484/Z/21/Z) and is supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (grant no. CC1002), the UK Medical Research Council (grant no. CC1002) and the Wellcome Trust (grant no. CC1002). M.B. is supported by an EMBO Postdoctoral Fellowship (grant no. ALTF 754-2023) and a Marie Skłodowska-Curie Actions via Horizon Europe Fellowship, underwritten through the EPSRC Horizon Europe Guarantee scheme. This research was funded in whole, or in part, by the Wellcome Trust (grant no. 224484/Z/21/Z, CC1002). For the purpose of Open Access, the authors have applied a Creative Commons Attribution (CC BY) public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Katharine Ullman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Coarse-grained simulations
-
Modelling techniques that simplify complex systems by grouping atoms or molecules into larger units (called beads), enabling the study of larger assemblies and longer timescales.
- Laminopathies
-
A group of rare genetic disorders caused by mutations in genes encoding nuclear lamina protein, leading to abnormalities in nuclear shape and structure and resulting in conditions such as progeria syndromes and muscular dystrophies.
- Midbody
-
A transient, protein-rich structure that assembles at the intercellular bridge during cytokinesis in mammalian cells.
- Open mitosis
-
A type of cell division in higher eukaryotes where the nuclear envelope completely breaks down to allow chromosome segregation.
- Stochastic optical reconstruction microscopy
-
A super-resolution imaging technique that uses the random switching of fluorescent molecules on and off to reconstruct highly detailed images beyond the diffraction limit.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Burigotto, M., Carlton, J.G. ESCRT-III function in membrane fission and repair. Nat Rev Mol Cell Biol (2025). https://doi.org/10.1038/s41580-025-00909-1
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41580-025-00909-1