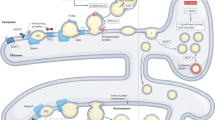
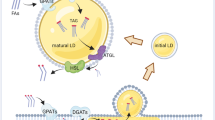
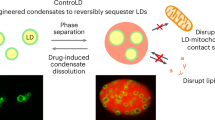
Abstract
Lipid droplets (LDs) are emerging as key factors in cellular physiology, with roles beyond energy storage, including metabolic homeostasis, signalling and development. Together with a growing list of functions, diverse LD populations are being identified in different tissue types as well as within the context of single cells. Here we summarize recent work highlighting LD diversity from three perspectives: their lipid and protein compositional heterogeneity; differences in abundance, size and spatial organization within cells; and the diverse contacts they form with other organelles, all of which contribute to LD function. We also discuss tools and approaches used to visualize LD heterogeneity, the role of LDs in pathophysiology and disease, and open questions in the field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Thiam, A. R., Farese, R. V. Jr. & Walther, T. C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786 (2013).
Wolk, M. & Fedorova, M. The lipid droplet lipidome. FEBS Lett. 598, 1215–1225 (2024).
Bersuker, K. et al. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev. Cell 44, 97–112.e117 (2018).
Krahmer, N. et al. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell Proteom. 12, 1115–1126 (2013).
Olzmann, J. A. & Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019).
Cohen, S. Lipid droplets as organelles. Int. Rev. Cell Mol. Biol. 337, 83–110 (2018).
Henne, W. M. The (social) lives, deaths, and biophysical phases of lipid droplets. Curr. Opin. Cell Biol. 82, 102178 (2023).
Petan, T., Jarc, E. & Jusovic, M. Lipid droplets in cancer: guardians of fat in a stressful world. Molecules 23, 1941 (2018).
Walther, T. C., Chung, J. & Farese, R. V. Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491–510 (2017).
Jackson, C. L. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 59, 88–96 (2019).
Zadoorian, A., Du, X. & Yang, H. Lipid droplet biogenesis and functions in health and disease. Nat. Rev. Endocrinol. 19, 443–459 (2023).
Farese, R. V. Jr. & Walther, T. C. Glycerolipid synthesis and lipid droplet formation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 15, a041246 (2023).
Bersuker, K. & Olzmann, J. A. Establishing the lipid droplet proteome: mechanisms of lipid droplet protein targeting and degradation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1166–1177 (2017).
Dhiman, R., Caesar, S., Thiam, A. R. & Schrul, B. Mechanisms of protein targeting to lipid droplets: a unified cell biological and biophysical perspective. Semin. Cell Dev. Biol. 108, 4–13 (2020).
Olarte, M. J., Swanson, J. M. J., Walther, T. C. & Farese, R. V. Jr. The CYTOLD and ERTOLD pathways for lipid droplet–protein targeting. Trends Biochem. Sci. 47, 39–51 (2022).
Choudhary, V., Ojha, N., Golden, A. & Prinz, W. A. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211, 261–271 (2015).
Jacquier, N. et al. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124, 2424–2437 (2011).
Cottier, S. & Schneiter, R. Lipid droplets form a network interconnected by the endoplasmic reticulum through which their proteins equilibrate. J. Cell Sci. 135, jcs258819 (2022).
Kumar, A., Yadav, S. & Choudhary, V. The evolving landscape of ER–LD contact sites. Front. Cell Dev. Biol. 12, 1483902 (2024).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017).
Wang, H. et al. Seipin is required for converting nascent to mature lipid droplets. eLife 5, e16582 (2016).
Salo, V. T. et al. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell 50, 478–493.e479 (2019).
Sui, X. et al. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217, 4080–4091 (2018).
Prasanna, X. et al. Seipin traps triacylglycerols to facilitate their nanoscale clustering in the endoplasmic reticulum membrane. PLoS Biol. 19, e3000998 (2021).
Zoni, V. et al. Seipin accumulates and traps diacylglycerols and triglycerides in its ring-like structure. Proc. Natl Acad. Sci. USA 118, e2017205118 (2021).
Renne, M. F., Corey, R. A., Ferreira, J. V., Stansfeld, P. J. & Carvalho, P. Seipin concentrates distinct neutral lipids via interactions with their acyl chain carboxyl esters. J. Cell Biol. 221, e202112068 (2022).
Wolins, N. E., Brasaemle, D. L. & Bickel, P. E. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580, 5484–5491 (2006).
Wilfling, F. et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 (2013).
Gao, Q. & Goodman, J. M. The lipid droplet — a well-connected organelle. Front. Cell Dev. Biol. 3, 49 (2015).
Kilwein, M. D. & Welte, M. A. Lipid droplet motility and organelle contacts. Contact https://doi.org/10.1177/2515256419895688 (2019).
Schuldiner, M. & Bohnert, M. A different kind of love — lipid droplet contact sites. Biochim. Biophys. Acta 1862, 1188–1196 (2017).
Herker, E., Vieyres, G., Beller, M., Krahmer, N. & Bohnert, M. Lipid droplet contact sites in health and disease. Trends Cell Biol. 31, 345–358 (2021).
Roberts, M. A. & Olzmann, J. A. Protein quality control and lipid droplet metabolism. Annu. Rev. Cell Dev. Biol. 36, 115–139 (2020).
Zechner, R., Madeo, F. & Kratky, D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18, 671–684 (2017).
Schott, M. B., Rozeveld, C. N., Weller, S. G. & McNiven, M. A. Lipophagy at a glance. J. Cell Sci. 135, jcs259402 (2022).
Barbosa, A. D. et al. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657 (2015).
Nguyen, T. B. et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e25 (2017).
Rambold, A. S., Cohen, S. & Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015).
Abela, G. S. Cholesterol crystals piercing the arterial plaque and intima trigger local and systemic inflammation. J. Clin. Lipidol. 4, 156–164 (2010).
Nguyen, T. B. & Olzmann, J. A. Lipid droplets and lipotoxicity during autophagy. Autophagy 13, 2002–2003 (2017).
Lee, H. et al. Cell cycle arrest induces lipid droplet formation and confers ferroptosis resistance. Nat. Commun. 15, 79 (2024).
Dierge, E. et al. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metab. 33, 1701–1715.e1705 (2021).
Lange, M. et al. FSP1-mediated lipid droplet quality control prevents neutral lipid peroxidation and ferroptosis. Nat. Cell Biol. 27, 1902–1913 (2025).
Papsdorf, K. et al. Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids. Nat. Cell Biol. 25, 672–684 (2023).
Li, Z. et al. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104–2113 (2012).
Mejhert, N. et al. Partitioning of MLX-family transcription factors to lipid droplets regulates metabolic gene expression. Mol. Cell 77, 1251–1264.e1259 (2020).
Orban, T., Palczewska, G. & Palczewski, K. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem. 286, 17248–17258 (2011).
Pan, H. et al. Centrins control chicken cone cell lipid droplet dynamics through lipid-droplet-localized SPDL1. Dev. Cell 58, 2528–2544.e2528 (2023).
Bosch, M. et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085 (2020).
Jarc, E. & Petan, T. A twist of FATe: lipid droplets and inflammatory lipid mediators. Biochimie 169, 69–87 (2020).
Chitraju, C. et al. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 53, 2141–2152 (2012).
Wang, L. et al. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature 581, 329–332 (2020).
Sui, X. et al. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature 581, 323–328 (2020).
Kim, S., Swanson, J. M. J. & Voth, G. A. Computational studies of lipid droplets. J. Phys. Chem. B 126, 2145–2154 (2022).
Dumesnil, C. et al. Cholesterol esters form supercooled lipid droplets whose nucleation is facilitated by triacylglycerols. Nat. Commun. 14, 915 (2023).
Rogers, S. et al. Triglyceride lipolysis triggers liquid crystalline phases in lipid droplets and alters the LD proteome. J. Cell Biol. 221, jcb202205053 (2022).
Mahamid, J. et al. Liquid–crystalline phase transitions in lipid droplets are related to cellular states and specific organelle association. Proc. Natl Acad. Sci. USA 116, 16866–16871 (2019).
Khor, V. K. et al. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS ONE 9, e105047 (2014).
Baumer, Y., Mehta, N. N., Dey, A. K., Powell-Wiley, T. M. & Boisvert, W. A. Cholesterol crystals and atherosclerosis. Eur. Heart J. 41, 2236–2239 (2020).
Turcu, A. F. & Auchus, R. J. Adrenal steroidogenesis and congenital adrenal hyperplasia. Endocrinol. Metab. Clin. North. Am. 44, 275–296 (2015).
Szkalisity, A. et al. Nuclear envelope-associated lipid droplets are enriched in cholesteryl esters and increase during inflammatory signaling. EMBO J. 44, 2774–2802 (2025).
Du, X. et al. ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 219, jcb201905162 (2020).
Krahmer, N. et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14, 504–515 (2011).
Aitchison, A. J., Arsenault, D. J. & Ridgway, N. D. Nuclear-localized CTP:phosphocholine cytidylyltransferase α regulates phosphatidylcholine synthesis required for lipid droplet biogenesis. Mol. Biol. Cell 26, 2927–2938 (2015).
Haider, A. et al. PCYT1A regulates phosphatidylcholine homeostasis from the inner nuclear membrane in response to membrane stored curvature elastic stress. Dev. Cell 45, 481–495.e488 (2018).
M’barek, K. B. et al. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604.e597 (2017).
Gok, M. O., Speer, N. O., Henne, W. M. & Friedman, J. R. ER-localized phosphatidylethanolamine synthase plays a conserved role in lipid droplet formation. Mol. Biol. Cell 33, ar11 (2022).
Kumar, S., Chitraju, C., Farese, R. V. Jr., Walther, T. C. & Burd, C. G. Conditional targeting of phosphatidylserine decarboxylase to lipid droplets. Biol. Open 10, bio058516 (2021).
Kurokawa, Y. et al. Microautophagy in the yeast vacuole depends on the activities of phosphatidylinositol 4-kinases, Stt4p and Pik1p. Biochim. Biophys. Acta Biomembr. 1862, 183416 (2020).
Bigay, J. & Antonny, B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell 23, 886–895 (2012).
Chorlay, A. & Thiam, A. R. Neutral lipids regulate amphipathic helix affinity for model lipid droplets. J. Cell Biol. 219, jcb201907099 (2020).
Ajjaji, D. et al. Dual binding motifs underpin the hierarchical association of perilipins1-3 with lipid droplets. Mol. Biol. Cell 30, 703–716 (2019).
Majchrzak, M. et al. Perilipin membrane integration determines lipid droplet heterogeneity in differentiating adipocytes. Cell Rep. 43, 114093 (2024).
Dias Araujo, A. R. et al. Surface tension-driven sorting of human perilipins on lipid droplets. J. Cell Biol. 223, jcb202403064 (2024).
Greenberg, A. S. et al. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266, 11341–11346 (1991).
Bulankina, A. V. et al. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185, 641–655 (2009).
Khaddaj, R., Stribny, J., Cottier, S. & Schneiter, R. Perilipin 3 promotes the formation of membrane domains enriched in diacylglycerol and lipid droplet biogenesis proteins. Front. Cell Dev. Biol. 11, 1116491 (2023).
Hsieh, K. et al. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125, 4067–4076 (2012).
Wang, H. & Sztalryd, C. Oxidative tissue: perilipin 5 links storage with the furnace. Trends Endocrinol. Metab. 22, 197–203 (2011).
Zhang, H. H. et al. Lipase-selective functional domains of perilipin a differentially regulate constitutive and protein kinase a-stimulated lipolysis. J. Biol. Chem. 278, 51535–51542 (2003).
Bi, J. et al. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J. Cell Sci. 125, 3568–3577 (2012).
Gao, Q. et al. Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J. Cell Biol. 216, 3199–3217 (2017).
Olarte, M. J. et al. Determinants of endoplasmic reticulum-to-lipid droplet protein targeting. Dev. Cell 54, 471–487.e477 (2020).
Henne, W. M., Reynolds, E. & Prinz, W. A. Lipid droplets: open questions and conceptual advances around a unique organelle. J. Cell Biol. 224, jcb202406019 (2025).
Windham, I. A. et al. APOE traffics to astrocyte lipid droplets and modulates triglyceride saturation and droplet size. J. Cell Biol. 223, jcb202305003 (2024).
Cartwright, B. R. & Goodman, J. M. Seipin: from human disease to molecular mechanism. J. Lipid Res. 53, 1042–1055 (2012).
Suzuki, M., Shinohara, Y., Ohsaki, Y. & Fujimoto, T. Lipid droplets: size matters. J. Electron Microsc. 60, S101–S116 (2011).
Calderon-Dominguez, M. et al. Fatty acid metabolism and the basis of brown adipose tissue function. Adipocyte 5, 98–118 (2016).
Wang, H. et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168 (2011).
Kim, Y. et al. Reorganization of mitochondria–organelle interactions during postnatal development in skeletal muscle. J. Physiol. 602, 891–912 (2024).
Kang, S. W. S. et al. A spatial map of hepatic mitochondria uncovers functional heterogeneity shaped by nutrient-sensing signaling. Nat. Commun. 15, 1799 (2024).
Eisenberg-Bord, M. et al. Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217, 269–282 (2018).
Teixeira, V. et al. Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217, 127–138 (2018).
Hariri, H. et al. Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 19, 57–72 (2018).
Alvarez-Guerra, I. et al. LDO proteins and Vac8 form a vacuole–lipid droplet contact site to enable starvation-induced lipophagy in yeast. Dev. Cell 59, 759–775.e755 (2024).
Hariri, H. et al. Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 218, 1319–1334 (2019).
Herms, A. et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6, 7176 (2015).
Pfisterer, S. G. et al. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat. Commun. 8, 14858 (2017).
Stephenson, R. A. et al. Sequestration to lipid droplets promotes histone availability by preventing turnover of excess histones. Development 148, dev199381 (2021).
Shai, N. et al. Systematic mapping of contact sites reveals tethers and a function for the peroxisome–mitochondria contact. Nat. Commun. 9, 1761 (2018).
Tashiro, S., Kakimoto, Y., Shinmyo, M., Fujimoto, S. & Tamura, Y. Improved split-GFP systems for visualizing organelle contact sites in yeast and human cells. Front. Cell Dev. Biol. 8, 571388 (2020).
Miner, G. E. et al. Contact-FP: a dimerization-dependent fluorescent protein toolkit for visualizing membrane contact site dynamics. Contact 7, 25152564241228911 (2024).
Li, X. et al. A fluorogenic complementation tool kit for interrogating lipid droplet–organelle interaction. J. Cell Biol. 223, e202311126 (2024).
Fung, H. K. H. et al. Genetically encoded multimeric tags for subcellular protein localization in cryo-EM. Nat. Methods 20, 1900–1908 (2023).
Binns, D. et al. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731 (2006).
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I. & Gaude, T. Peroxisome extensions deliver the arabidopsis SDP1 lipase to oil bodies. Proc. Natl Acad. Sci. USA 112, 4158–4163 (2015).
Eisenberg-Bord, M., Shai, N., Schuldiner, M. & Bohnert, M. A tether is a tether is a tether: tethering at membrane contact sites. Dev. Cell 39, 395–409 (2016).
Knedlik, T. & Giacomello, M. Temporal dynamics of membrane contact sites. Nat. Cell Biol. 26, 1822–1824 (2024).
Castro, I. G. et al. Systematic analysis of membrane contact sites in Saccharomyces cerevisiae uncovers modulators of cellular lipid distribution. eLife 11, e74602 (2022).
Arlt, H. et al. Seipin forms a flexible cage at lipid droplet formation sites. Nat. Struct. Mol. Biol. 29, 194–202 (2022).
Yan, R. et al. Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256.e244 (2018).
Song, J. et al. Identification of two pathways mediating protein targeting from ER to lipid droplets. Nat. Cell Biol. 24, 1364–1377 (2022).
Malis, Y. et al. Rab1b facilitates lipid droplet growth by ER-to-lipid droplet targeting of DGAT2. Sci. Adv. 10, eade7753 (2024).
Xu, D. et al. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995 (2018).
Ozeki, S. et al. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118, 2601–2611 (2005).
Li, D. et al. The ER-localized protein DFCP1 modulates ER–lipid droplet contact formation. Cell Rep. 27, 343–358.e345 (2019).
Jayson, C. B. K. et al. Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol. Biol. Cell 29, 2045–2054 (2018).
Soni, K. G. et al. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122, 1834–1841 (2009).
Wilfling, F. et al. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3, e01607 (2014).
Chen, S. et al. VPS13A and VPS13C influence lipid droplet abundance. Contact 5, 25152564221125613 (2022).
Kumar, N. et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018).
Xu, N. et al. The FATP1–DGAT2 complex facilitates lipid droplet expansion at the ER–lipid droplet interface. J. Cell Biol. 198, 895–911 (2012).
de la Rosa Rodriguez, M. A. et al. Hypoxia-inducible lipid droplet-associated induces DGAT1 and promotes lipid storage in hepatocytes. Mol. Metab. 47, 101168 (2021).
Datta, S., Liu, Y., Hariri, H., Bowerman, J. & Henne, W. M. Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER–droplet contacts. J. Cell Biol. 218, 1335–1351 (2019).
Zouiouich, M. et al. MOSPD2 is an endoplasmic reticulum–lipid droplet tether functioning in LD homeostasis. J. Cell Biol. 221, e202110044 (2022).
Joshi, A. S., Ragusa, J. V., Prinz, W. A. & Cohen, S. Multiple C2 domain-containing transmembrane proteins promote lipid droplet biogenesis and growth at specialized endoplasmic reticulum subdomains. Mol. Biol. Cell 32, 1147–1157 (2021).
Mizrak, A. et al. Single-molecule analysis of protein targeting from the endoplasmic reticulum to lipid droplets. Preprint at bioRxiv https://doi.org/10.1101/2024.08.27.610018 (2024).
Markgraf, D. F. et al. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep. 6, 44–55 (2014).
Bem, D. et al. Loss-of-function mutations in RAB18 cause warburg micro syndrome. Am. J. Hum. Genet. 88, 499–507 (2011).
Carpanini, S. M. et al. A novel mouse model of warburg micro syndrome reveals roles for RAB18 in eye development and organisation of the neuronal cytoskeleton. Dis. Model. Mech. 7, 711–722 (2014).
Martin, S., Driessen, K., Nixon, S. J., Zerial, M. & Parton, R. G. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280, 42325–42335 (2005).
Xu, M. et al. Rab2A-mediated Golgi–lipid droplet interactions support very-low-density lipoprotein secretion in hepatocytes. EMBO J. 43, 6383–6409 (2024).
Tang, S. et al. Association of HSD17B13 rs72613567: TA allelic variant with liver disease: review and meta-analysis. BMC Gastroenterol. 21, 490 (2021).
Du, Y. et al. A possible role of VPS13B in the formation of golgi-lipid droplet contacts associating with the ER. Contact 6, 25152564231195718 (2023).
Toulmay, A. et al. Vps13-like proteins provide phosphatidylethanolamine for GPI anchor synthesis in the ER. J. Cell Biol. 221, e202111095 (2022).
Neuman, S. D., Levine, T. P. & Bashirullah, A. A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol. 32, 962–974 (2022).
Ugrankar, R. et al. Drosophila snazarus regulates a lipid droplet population at plasma membrane-droplet contacts in adipocytes. Dev. Cell 50, 557–572.e555 (2019).
Matthaeus, C. et al. EHD2-mediated restriction of caveolar dynamics regulates cellular fatty acid uptake. Proc. Natl Acad. Sci. USA 117, 7471–7481 (2020).
Kimmel, A. R. & Sztalryd, C. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu. Rev. Nutr. 36, 471–509 (2016).
Kuramoto, K. et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 287, 23852–23863 (2012).
Wang, H. et al. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J. Lipid Res. 54, 953–965 (2013).
Benador, I. Y. et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885 e866 (2018).
Bosma, M. et al. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. Biochim. Biophys. Acta 1831, 844–852 (2013).
Keenan, S. N. et al. Perilipin 5 deletion in hepatocytes remodels lipid metabolism and causes hepatic insulin resistance in mice. Diabetes 68, 543–555 (2019).
Gallardo-Montejano, V. I. et al. Perilipin 5 links mitochondrial uncoupled respiration in brown fat to healthy white fat remodeling and systemic glucose tolerance. Nat. Commun. 12, 3320 (2021).
Benador, I. Y., Veliova, M., Liesa, M. & Shirihai, O. S. Mitochondria bound to lipid droplets: where mitochondrial dynamics regulate lipid storage and utilization. Cell Metab. 29, 827–835 (2019).
Pollak, N. M. et al. The interplay of protein kinase a and perilipin 5 regulates cardiac lipolysis. J. Biol. Chem. 290, 1295–1306 (2015).
Miner, G. E. et al. PLIN5 interacts with FATP4 at membrane contact sites to promote lipid droplet-to-mitochondria fatty acid transport. Dev. Cell 58, 1250–1265.e1256 (2023).
Ouyang, Q. et al. Rab8a as a mitochondrial receptor for lipid droplets in skeletal muscle. Dev. Cell 58, 289–305.e286 (2023).
Kang, S. W. S. et al. Spatially resolved rewiring of mitochondria-lipid droplet interactions in hepatic lipid homeostasis. Preprint at bioRxiv https://doi.org/10.1101/2024.12.10.627730 (2024).
Jagerstrom, S. et al. Lipid droplets interact with mitochondria using SNAP23. Cell Biol. Int. 33, 934–940 (2009).
Young, P. A. et al. Long-chain acyl-CoA synthetase 1 interacts with key proteins that activate and direct fatty acids into niche hepatic pathways. J. Biol. Chem. 293, 16724–16740 (2018).
Wang, J. et al. An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D–TSG101 interactions. Nat. Commun. 12, 1252 (2021).
Vietri, M., Radulovic, M. & Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020).
Freyre, C. A. C., Rauher, P. C., Ejsing, C. S. & Klemm, R. W. MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell 76, 811–825.e814 (2019).
Hong, Z. et al. Mitoguardin-2-mediated lipid transfer preserves mitochondrial morphology and lipid droplet formation. J. Cell Biol. 221, e202207022 (2022).
Kim, H., Lee, S., Jun, Y. & Lee, C. Structural basis for mitoguardin-2 mediated lipid transport at ER–mitochondrial membrane contact sites. Nat. Commun. 13, 3702 (2022).
Wanders, R. J. A., Baes, M., Ribeiro, D., Ferdinandusse, S. & Waterham, H. R. The physiological functions of human peroxisomes. Physiol. Rev. 103, 957–1024 (2023).
Amado, L. et al. Pex3 promotes formation of peroxisome-peroxisome and peroxisome–lipid droplet contact sites. Sci. Rep. 15, 24480 (2025).
Traver, M. S. & Bartel, B. The ubiquitin–protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization. Proc. Natl Acad. Sci. USA 120, e2304870120 (2023).
Chang, C. L. et al. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J. Cell Biol. 218, 2583–2599 (2019).
Zimmermann, J. A. et al. Functional multi-organelle units control inflammatory lipid metabolism of macrophages. Nat. Cell Biol. 26, 1261–1273 (2024).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Schott, M. B. et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 218, 3320–3335 (2019).
Kaushik, S. & Cuervo, A. M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015).
Kaushik, S. & Cuervo, A. M. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 12, 432–438 (2016).
Kaushik, S. et al. Chaperone-mediated autophagy regulates adipocyte differentiation. Sci. Adv. 8, eabq2733 (2022).
Khawaja, R. R. et al. Sex-specific and cell-type-specific changes in chaperone-mediated autophagy across tissues during aging. Nat. Aging 5, 691–708 (2025).
Haemmerle, G. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006).
Paar, M. et al. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J. Biol. Chem. 287, 11164–11173 (2012).
Schott, M. B. et al. β-Adrenergic induction of lipolysis in hepatocytes is inhibited by ethanol exposure. J. Biol. Chem. 292, 11815–11828 (2017).
Vargas, J. N. S., Hamasaki, M., Kawabata, T., Youle, R. J. & Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 24, 167–185 (2023).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Chung, J. et al. The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets. Nat. Cell Biol. 25, 1101–1110 (2023).
Wan, N. et al. Spartin-mediated lipid transfer facilitates lipid droplet turnover. Proc. Natl Acad. Sci. USA 121, e2314093121 (2024).
Schroeder, B. et al. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907 (2015).
Schulze, R. J. et al. Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol. Commun. 1, 140–152 (2017).
Lizaso, A., Tan, K. T. & Lee, Y. H. β-Adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243 (2013).
Li, Z. et al. A novel Rab10–EHBP1–EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2, e1601470 (2016).
Patel, H. et al. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat. Genet. 31, 347–348 (2002).
Ralhan, I., Chang, C. L., Lippincott-Schwartz, J. & Ioannou, M. S. Lipid droplets in the nervous system. J. Cell Biol. 220, e202102136 (2021).
Ouimet, M. et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 13, 655–667 (2011).
van Zutphen, T. et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301 (2014).
Wang, C. W., Miao, Y. H. & Chang, Y. S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206, 357–366 (2014).
Vevea, J. D. et al. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 (2015).
Toulmay, A. & Prinz, W. A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202, 35–44 (2013).
Seo, A. Y. et al. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. eLife 6, e21690 (2017).
Schulze, R. J. et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl Acad. Sci. USA 117, 32443–32452 (2020).
Menon, D. et al. ARL8B mediates lipid droplet contact and delivery to lysosomes for lipid remobilization. Cell Rep. 42, 113203 (2023).
Windham, I. A. & Cohen, S. Lipid droplets go through a (liquid crystalline) phase. J. Cell Biol. 221, e202210008 (2022).
Soltysik, K. et al. Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220, e202005026 (2021).
Romanauska, A. & Kohler, A. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell 174, 700–715.e718 (2018).
Wang, C. W., Miao, Y. H. & Chang, Y. S. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127, 1214–1228 (2014).
Grippa, A. et al. The seipin complex Fld1/Ldb16 stabilizes ER–lipid droplet contact sites. J. Cell Biol. 211, 829–844 (2015).
Castro, I. G. et al. Promethin is a conserved seipin partner protein. Cells 8, 268 (2019).
Chung, J. et al. LDAF1 and seipin form a lipid droplet assembly complex. Dev. Cell 51, 551–563.e557 (2019).
Henne, W. M. et al. Mdm1/Snx13 is a novel ER–endolysosomal interorganelle tethering protein. J. Cell Biol. 210, 541–551 (2015).
Joshi, A. S. et al. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9, 2940 (2018).
Guyard, V. et al. ORP5 and ORP8 orchestrate lipid droplet biogenesis and maintenance at ER–mitochondria contact sites. J. Cell Biol. 221, e202112107 (2022).
Monks, J. et al. Perilipin-2 promotes lipid droplet-plasma membrane interactions that facilitate apocrine lipid secretion in secretory epithelial cells of the mouse mammary gland. Front. Cell Dev. Biol. 10, 958566 (2022).
Boutant, M. et al. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36, 1543–1558 (2017).
Brasaemle, D. L. & Wolins, N. E. Isolation of lipid droplets from cells by density gradient centrifugation. Curr. Protoc. Cell Biol. 72, 3.15.11–13.15.13 (2016).
Zhang, S. et al. Morphologically and functionally distinct lipid droplet subpopulations. Sci. Rep. 6, 29539 (2016).
Wolins, N. E. et al. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280, 19146–19155 (2005).
Speer, N. O. et al. Tld1 is a regulator of triglyceride lipolysis that demarcates a lipid droplet subpopulation. J. Cell Biol. 223, e202303026 (2024).
Shimobayashi, S. F. & Ohsaki, Y. Universal phase behaviors of intracellular lipid droplets. Proc. Natl Acad. Sci. USA 116, 25440–25445 (2019).
Zanellati, M. C., Hsu, C. H. & Cohen, S. Imaging interorganelle contacts at a glance. J. Cell Sci. 137, e262020 (2024).
Gamuyao, R. & Chang, C. L. Imaging and proteomics toolkits for studying organelle contact sites. Front. Cell Dev. Biol. 12, 1466915 (2024).
Herms, A. et al. Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr. Biol. 23, 1489–1496 (2013).
Rao, M. J. & Goodman, J. M. Seipin: harvesting fat and keeping adipocytes healthy. Trends Cell Biol. 31, 912–923 (2021).
Sheka, A. C. et al. Nonalcoholic steatohepatitis: a review. JAMA 323, 1175–1183 (2020).
Mitrofanova, A., Merscher, S. & Fornoni, A. Kidney lipid dysmetabolism and lipid droplet accumulation in chronic kidney disease. Nat. Rev. Nephrol. 19, 629–645 (2023).
Wagner, R. et al. Metabolic implications of pancreatic fat accumulation. Nat. Rev. Endocrinol. 18, 43–54 (2022).
Goldberg, I. J., Trent, C. M. & Schulze, P. C. Lipid metabolism and toxicity in the heart. Cell Metab. 15, 805–812 (2012).
Cruz, A. L. S., Barreto, E. A., Fazolini, N. P. B., Viola, J. P. B. & Bozza, P. T. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 11, 105 (2020).
Alzheimer, A., Stelzmann, R. A., Schnitzlein, H. N. & Murtagh, F. R. An english translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde”. Clin. Anat. 8, 429–431 (1995).
Haney, M. S. et al. APOE4/4 is linked to damaging lipid droplets in Alzheimer’s disease microglia. Nature 628, 154–161 (2024).
Prakash, P. et al. Amyloid-β induces lipid droplet-mediated microglial dysfunction via the enzyme DGAT2 in Alzheimer’s disease. Immunity 58, 1536–1552.e8 (2025).
Herker, E. Lipid droplets in virus replication. FEBS Lett. 598, 1299–1300 (2024).
Acknowledgements
We thank Y. Malis for assistance with figures and with research for Table 1. We also thank D. Nicastro and L. Gui for assistance with the cryo-electron tomography images used in Fig. 2. The authors are supported by funding from the National Institutes of Health under awards R35GM119768 and DK126887 (W.M.H.) and R35GM133460 (S.C.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Birefringence
-
An optical feature of some materials that allows polarized light to have different refractive indices when it passes through the material, revealing a sometimes rainbow-like diffraction pattern to the polarized light. When used in optical imaging of lipid droplets, smectic liquid–crystalline lipid droplets display a distinctive Maltese cross pattern.
- BODIPY
-
Refers to several fluorescent dyes 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene that partition inside lipid droplets and are used for lipid droplet labelling.
- Fluorogen-activated bimolecular complementation
-
A microscopy technique used in studying organelle contacts where non-fluorescent protein segments are fused to proteins of interest localized on different organelles. When these proteins come together the non-fluorescent segments dimerize, forming a docking site for a fluorescent organic dye molecule that then labels the interaction site.
- Glucose exhaustion
-
A metabolic condition characterized by the depletion of available glucose for cells to utilize in metabolism.
- Macrophage foam cells
-
A pathogenic cell type that accumulates in cardiovascular disease when macrophage cells absorb excess LDL particles and accumulate intracellular lipid droplets rich in cholesteryl esters.
- NRZ–SNARE–RAB18
-
A tethering complex mediated by RAB18 at the lipid droplet interacting with the endoplasmic reticulum-localized NAG–RINT1–ZW10 (NRZ) complex and their associated SNAREs.
- Phase separation
-
The process where a mixture demixes into two distinct liquid phases, driven by interactions between molecules.
- Smectic
-
A phase of a liquid crystal where molecules are arranged in layers.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Henne, W.M., Cohen, S. Heterogeneity, dynamics and organelle interactions of lipid droplets. Nat Rev Mol Cell Biol (2026). https://doi.org/10.1038/s41580-025-00945-x
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41580-025-00945-x