Abstract
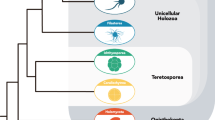
All animals develop from a single-celled zygote into a complex multicellular organism through a series of precisely orchestrated processes1,2. Despite the remarkable conservation of early embryogenesis across animals, the evolutionary origins of how and when this process first emerged remain elusive. Here, by combining time-resolved imaging and transcriptomic profiling, we show that single cells of the ichthyosporean Chromosphaera perkinsii—a close relative that diverged from animals about 1 billion years ago3,4—undergo symmetry breaking and develop through cleavage divisions to produce a prolonged multicellular colony with distinct co-existing cell types. Our findings about the autonomous and palintomic developmental program of C. perkinsii hint that such multicellular development either is much older than previously thought or evolved convergently in ichthyosporeans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used for quantifications and Supplementary Videos 1–10 are available at Figshare (https://figshare.com/s/f20f6d471c719990471c)92. For all other raw images required, they are available on request due to their large data sizes. Raw RNA-seq data are publicly available on https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1091032.
Code availability
All the generated code for transcriptomic analysis is available at Zenodo (https://doi.org/10.5281/zenodo.13352464)93.
References
Kalinka, A. T. & Tomancak, P. The evolution of early animal embryos: conservation or divergence? Trends Ecol. Evol. 27, 385–393 (2012).
Salazar-Ciudad, I. Morphological evolution and embryonic developmental diversity in metazoa. Development 137, 531–539 (2010).
Grau-Bové, X. et al. Dynamics of genomic innovation in the unicellular ancestry of animals. eLife 6, e26036 (2017).
Ocaña-Pallarès, E. et al. Divergent genomic trajectories predate the origin of animals and fungi. Nature 609, 747–753 (2022).
Sebé-Pedrós, A., Degnan, B. M. & Ruiz-Trillo, I. The origin of Metazoa: a unicellular perspective. Nat. Rev. Genet. https://doi.org/10.1038/nrg.2017.21 (2017).
Brunet, T. & King, N. in The Evolution of Multicellularity (eds Herron, M. D. et al.) Ch. 13 (CRC, 2022).
Knoll, A. H. The multiple origins of complex multicellularity. Annu. Rev. Earth Planet Sci. 39, 217–239 (2011).
De Smet, I. & Beeckman, T. Asymmetric cell division in land plants and algae: the driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12, 177–188 (2011).
Gönczy, P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355–366 (2008).
Lechler, T. & Mapelli, M. Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat. Rev. Mol. Cell Biol. 22, 691–708 (2021).
Tsai, T. Y. C., Garner, R. M. & Megason, S. G. Adhesion-based self-organization in tissue patterning. Annu. Rev. Cell Dev. Biol. 38, 349–374 (2022).
Wu, D., Yamada, K. M. & Wang, S. Tissue morphogenesis through dynamic cell and matrix interactions. Annu. Rev. Cell Dev. Biol. 39, 123–144 (2023).
Arendt, D. et al. The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016).
Bell, G. & Mooers, A. O. Size and complexity among multicellular organisms. Biol. J. Linn. Soc. 60, 345–363 (1997).
Márquez-Zacarías, P. et al. Evolution of cellular differentiation: from hypotheses to models. Trends Ecol. Evol. 36, 49–60 (2021).
Wolpert, L. Do we understand development? Science (1979) 266, 571–572 (1994).
Ruiz-Trillo, I. & de Mendoza, A. Towards understanding the origin of animal development. Development https://doi.org/10.1242/dev.192575 (2020).
Brunet, T. et al. A flagellate-to-amoeboid switch in the closest living relatives of animals. eLife 10, 1–73 (2021).
Brunet, T. et al. Light-regulated collective contractility in a multicellular choanoflagellate. Science https://doi.org/10.1126/science.aay2346 (2019).
Fairclough, S. R., Dayel, M. J. & King, N. Multicellular development in a choanoflagellate. Curr. Biol. https://doi.org/10.1016/j.cub.2010.09.014 (2010).
Parra-Acero, H. et al. Integrin-mediated adhesion in the unicellular holozoan Capsaspora owczarzaki. Curr. Biol. https://doi.org/10.1016/j.cub.2020.08.015 (2020).
Sebé-Pedrós, A. et al. Regulated aggregative multicellularity in a close unicellular relative of metazoa. eLife https://doi.org/10.7554/eLife.01287 (2013).
Pérez-Posada, A., Dudin, O., Ocaña-Pallarès, E., Ruiz-Trillo, I. & Ondracka, A. Cell cycle transcriptomics of Capsaspora provides insights into the evolution of cyclin-CDK machinery. PLoS Genet. https://doi.org/10.1371/journal.pgen.1008584 (2020).
Xiao, S. et al. The Weng’an biota and the Ediacaran radiation of multicellular eukaryotes. Natl Sci. Rev. 1, 498–520 (2014).
Ros-Rocher, N. et al. Chemical factors induce aggregative multicellularity in a close unicellular relative of animals. Proc. Natl Acad. Sci. USA 120, e2216668120 (2023).
Woznica, A. et al. Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1605015113 (2016).
Levin, T. C., Greaney, A. J., Wetzel, L. & King, N. The Rosetteless gene controls development in the choanoflagellate S. rosetta. eLife https://doi.org/10.7554/eLife.04070 (2014).
Levin, T. C. & King, N. Evidence for sex and recombination in the Choanoflagellate Salpingoeca rosetta. Curr. Biol. https://doi.org/10.1016/j.cub.2013.08.061 (2013).
Dayel, M. J. et al. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82 (2011).
Laundon, D., Larson, B. T., McDonald, K., King, N. & Burkhardt, P. The architecture of cell differentiation in choanoflagellates and sponge choanocytes. PLoS Biol. 17, e3000226 (2019).
Phillips, J. E., Santos, M., Konchwala, M., Xing, C. & Pan, D. Genome editing in the unicellular holozoan Capsaspora owczarzaki suggests a premetazoan role for the Hippo pathway in multicellular morphogenesis. eLife 11, e77598 (2022).
Glockling, S. L., Marshall, W. L. & Gleason, F. H. Phylogenetic interpretations and ecological potentials of the Mesomycetozoea (Ichthyosporea). Fungal Ecol. 6, 237–247 (2013).
Mendoza, L., Taylor, J. W. & Ajello, L. The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu. Rev. Microbiol. 56, 315–344 (2002).
Ondracka, A., Dudin, O. & Ruiz-Trillo, I. Decoupling of nuclear division cycles and cell size during the coenocytic growth of the Ichthyosporean Sphaeroforma arctica. Curr. Biol. 28, 1964–1969.e2 (2018).
Dudin, O., Wielgoss, S., New, A. M. & Ruiz-Trillo, I. Regulation of sedimentation rate shapes the evolution of multicellularity in a close unicellular relative of animals. PLoS Biol. 20, e3001551 (2022).
Dudin, O. et al. A unicellular relative of animals generates a layer of polarized cells by actomyosin-dependent cellularization. eLife 8, e49801 (2019).
McCartney, B. & Dudin, O. Cellularization across eukaryotes: conserved mechanisms and novel strategies. Curr. Opin. Cell Biol. 80, 102157 (2023).
Shah, H. et al. Life-cycle-coupled evolution of mitosis in close relatives of animals. Nature https://doi.org/10.1038/s41586-024-07430-z (2024).
Olivetta, M. & Dudin, O. The nuclear-to-cytoplasmic ratio drives cellularization in the close animal relative Sphaeroforma arctica. Curr. Biol. 33, 1597–1605.e3 (2023).
Vasudevan, S., Seli, E. & Steitz, J. A. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev. 20, 138–146 (2006).
Levin, M. et al. The mid-developmental transition and the evolution of animal body plans. Nature https://doi.org/10.1038/nature16994 (2016).
Carvalho-Santos, Z., Azimzadeh, J., Pereira-Leal, J. B. & Bettencourt-Dias, M. Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 (2011).
Brunet, T. & Booth, D. S. Cell polarity in the protist-to-animal transition. Curr. Top. Dev. Biol. 154, 1–36 (2023).
Wright, B. A., Kvansakul, M., Schierwater, B. & Humbert, P. O. Cell polarity signalling at the birth of multicellularity: what can we learn from the first animals. Front. Cell. Dev. Biol. 10, 1024489 (2022).
Klapholz, B. & Brown, N. H. Talin - the master of integrin adhesions. J. Cell Sci. 130, 2435–2446 (2017).
Bays, J. L. & DeMali, K. A. Vinculin in cell–cell and cell–matrix adhesions. Cell. Mol. Life Sci. 74, 2999–3009 (2017).
Sebé-Pedrós, A., De Mendoza, A., Lang, B. F., Degnan, B. M. & Ruiz-Trillo, I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan capsaspora owczarzaki. Mol. Biol. Evol. https://doi.org/10.1093/molbev/msq309 (2011).
De Mendoza, A. et al. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1311818110 (2013).
Serra, D. et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019).
Palmer, A. R. Symmetry breaking and the evolution of development. Science 306, 828–833 (2004).
Brunet, T. & King, N. The origin of animal multicellularity and cell differentiation. Dev. Cell https://doi.org/10.1016/j.devcel.2017.09.016 (2017).
Raghu-kumar, S. Occurrence of the Thraustochytrid, Corallochytrium limacisporum gen. et sp. nov. in the coral reef lagoons of the Lakshadweep Islands in the Arabian Sea. Botanica Marina 30, 83–90 (1987).
Hehenberger, E. et al. Novel predators reshape holozoan phylogeny and reveal the presence of a two-component signaling system in the ancestor of animals. Curr. Biol. 27, 2043–2050.e6 (2017).
Ros-Rocher, N., Pérez-Posada, A., Leger, M. M. & Ruiz-Trillo, I. The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition. Open Biol. 11, 200359 (2021).
Larson, B. T. et al. Biophysical principles of choanoflagellate self-organization. Proc. Natl Acad. Sci. USA 117, 1303–1311 (2020).
Yang, C. et al. The tempo of Ediacaran evolution. Sci. Adv. 7, 9643 (2021).
Cunningham, J. A., Vargas, K., Yin, Z., Bengtson, S. & Donoghue, P. C. J. The Weng’an Biota (Doushantuo Formation): an Ediacaran window on soft-bodied and multicellular microorganisms. J. Geol. Soc. London 174, 793–802 (2017).
Yin, Z. et al. Nuclei and nucleoli in embryo-like fossils from the Ediacaran Weng’an Biota. Precambrian Res. 301, 145–151 (2017).
Yin, Z. et al. Developmental biology of Helicoforamina reveals holozoan affinity, cryptic diversity, and adaptation to heterogeneous environments in the early Ediacaran Weng’an biota (Doushantuo Formation, South China). Sci. Adv. 6, 83–95 (2020).
Sun, W., Yin, Z., Liu, P., Zhu, M. & Donoghue, P. Developmental biology of Spiralicellula and the Ediacaran origin of crown metazoans. Proc. R. Soc. Lond. B Biol. Sci. 291, 20240101 (2024).
Yin, Z. et al. The early Ediacaran Caveasphaera foreshadows the evolutionary origin of animal-like embryology. Curr. Biol. 29, 4307–4314.e2 (2019).
Yin, Z. et al. Diverse and complex developmental mechanisms of early Ediacaran embryo-like fossils from the Weng’an Biota, southwest China. Phil. Trans. R. Soc. Lond. B Biol. Sci. 377, 20210032 (2022).
Xiao, S. Mitotic topologies and mechanics of Neoproterozoic algae and animal embryos. Paleobiology 28, 244–250 (2002).
Shuhai, X. et al. Fossil preservation in the Neoproterozoic Doushantuo phosphorite Lagerstätte, South China. Lethaia 32, 219–238 (1999).
Chen, L., Xiao, S., Pang, K., Zhou, C. & Yuan, X. Cell differentiation and germ-soma separation in Ediacaran animal embryo-like fossils. Nature 516, 238–241 (2014).
Huldtgren, T. et al. Fossilized nuclei and germination structures identify ediacaran ‘animal embryos’ as encysting protists. Science 334, 1696–1699 (2011).
Alexa, A. & Rahnenfuhrer, J. topGO: enrichment analysis for gene ontology. R package version 2.42.0 https://rdrr.io/bioc/topGO/ (2020).
Gambarotto, D. et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2018).
Machado, S., Mercier, V. & Chiaruttini, N. LimeSeg: a coarse-grained lipid membrane simulation for 3D image segmentation. BMC Bioinf. 20, 1–12 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Multicellgenome Lab. Genome - Chromosphaera perkinsii. Figshare https://figshare.com/articles/dataset/Genome_-_Chromosphaera_perkinsii/5426494 (2024).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2016).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288 (2012).
Chen, Y., Lun, A. T. L. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 5, 1438 (2016).
Meyer, D. et al. e1071: misc functions of the Department of Statistics, Probability Theory Group (formerly: E1071), TU Wien. R package version 1.7-15 https://rdrr.io/rforge/e1071/ (2024).
Garnier, S. et al. viridis: colorblind-friendly color maps for R. R package version 0.6.4 https://rdrr.io/cran/viridis/ (2024).
Barter, R. & Yu, B. superheat: a graphical tool for exploring complex datasets using heatmaps. R package version 0.1.0 https://rdrr.io/cran/superheat/ (2019).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinf. 10, 1–9 (2009).
Boutet, E. et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54 (2016).
Rost, B. Enzyme function less conserved than anticipated. J. Mol. Biol. 318, 595–608 (2002).
Burns, J. A., Paasch, A., Narechania, A. & Kim, E. Comparative genomics of a bacterivorous green alga reveals evolutionary causalities and consequences of phago-mixotrophic mode of nutrition. Genome Biol. Evol. 7, 3047–3061 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Haas, B. J. et al. TransDecoder source. GitHub https://github.com/TransDecoder/TransDecoder (2024).
Engel, S. R. et al. New data and collaborations at the Saccharomyces Genome Database: updated reference genome, alleles, and the Alliance of Genome Resources. Genetics 220, iyab224 (2022).
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880 (2002).
Eichinger, I. et al. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 (2005).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Wei, T. & Simko, V. corrplot: visualization of a correlation matrix. R package version 0.92. https://rdocumentation.org/packages/corrplot/versions/0.94 (2024).
Gearty, W. & Jones, L. A. rphylopic: an R package for fetching, transforming, and visualising PhyloPic silhouettes. Methods Ecol. Evol. 14, 2700–2708 (2023).
Olivetta, M. et al. Source data for ‘A multicellular developmental program in a close animal relative’. Figshare https://figshare.com/s/f20f6d471c719990471c (2024).
Olivetta, M. et al. Code for ‘A multicellular developmental program in a close animal relative’. Zenodo https://doi.org/10.5281/zenodo.13352464 (2024).
Acknowledgements
We thank P. Gönczy, G. Dey, A. de Mendoza, C. Martín-Durán, H. Shah and A. Woglar for comments on the manuscript and general feedback; H. Suga for C. perkinsii cultures; and both the EPFL Bio Imaging and Optics Platform and the EPFL Gene Expression Core Facility for their support. We acknowledge funding from US National Science Foundation grant no. OIA-1826734 (J.B.), Swiss National Science Foundation Ambizione grant no. PZ00P3_185859 (O.D. and M.O.) and Swiss National Science Foundation Starting grant no. TMSGI3_218007 (O.D. and M.O.).
Author information
Authors and Affiliations
Contributions
Conceptualization: O.D. Methodology: M.O., C.B., N.C., J.B., O.D. Investigation: M.O., C.B., N.C., J.B. and O.D. Visualization: M.O., N.C., J.B. and O.D. Funding acquisition: J.B. and O.D. Project administration: O.D. Supervision: O.D. Writing—original draft: O.D. Writing—review and editing: M.O., N.C., J.B. and O.D.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Philip Donoghue and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Culture synchronization and growth dynamics of C. perkinsii.
(a) Schematic of the synchronization protocol of C. perkinsii. (b) Distribution of cells undergoing their first cleavage division or cell release throughout the life cycle at 23 °C, following either synchronization or not (n non-synchronized=129 cell/colony, n synchronized=233 cell/colony). Data are mean ± s.d. (c) The average time at which non-synchronized and synchronized cells undergo their 1st cleavage in a bulk culture (n non-synchronized=129 cell/colony, n synchronized=233 cell/colony). Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value. (d) Percentage of free-swimming flagellated cells over time (n 0 = 112 cells, n 24 = 84 cells, n 44 = 110 cells, n46 = 70 cells, n51 = 99 cells, n53 = 73 cells, n68 = 116 cells, n70 = 105 cells, n72 = 68 cells, n76 = 77 cells, n99 = 104 cells, n120 = 67 cells). Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value. (e) Cell diameter over time of 10 single cell traces aligned to time. Variability increases from 75 hr onwards due to asynchronous cell release. Data are mean ± s.d. (f) Cell diameter over time of fixed cells (n = 1571 cell/colony). The colours represent 3 independent replicates. (g) Distribution of number of nuclei per cell/colony across the life-cycle (n = 1571 cell/colony). The colours represent 3 independent replicates. Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value.
Extended Data Fig. 2 Transcriptional dynamics in C. perkinsii’s palintomic lifecycle and comparative analysis with S. arctica and early-branching animals.
(a) Multiple dimensional scaling (MDS) plot of RNAseq counts of 3 independent developmental time courses of C. perkinsii. “B” replicates were excluded from further analyses due to evidence of infection (Extended Data Table 1). (b) Expression profile distributions for the 5 distinct gene expression clusters for both replicates. Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value. (c) Heatmaps of gene expression dynamics between C. perkinsii and S. arctica highlight distinct transcriptional programs. (d) Statistical evaluation of gene expression correlation between C. perkinsii and S. arctica in select GO categories. (e) Correlation values comparing C. perkinsii to S. arctica expression patterns for genes important during coenocytic development of S. arctica35. (f) Network strategy used to decouple distinct expression patterns occurring within an orthogroup (methods). (g) Average correlation of OG expression values between early diverging animals and C. perkinsii. Orthogroups with an average correlation value greater than 0.5 (blue line) were subjected to GO enrichment analyses (Supplementary Tables 1, 2). (h) Average correlation of OG expression values between all animals and C. perkinsii. OGs with an average correlation value greater than 0.25 (blue line) were subjected to GO enrichment analyses (Supplementary Tables 1, 2). (i) Distribution of gene expression correlation values between C. perkinsii and each animal for non-developmental genes (Control Not DE) and differentially expressed gene clusters. The white dashed line indicates the median correlation value of the control distribution. (j) Average gene expression correlation among all animals plus C. perkinsii for a random sample of 34 OGs (black bars), developmental genes (pink line), and glycolysis genes (not developmentally regulated; blue line). (k) Pairwise correlation coefficient plots of gene clusters showing pairwise correlations among all animals and C. perkinsii. (l) Heatmap of gene expression for select OGs between C. perkinsii and developmental stages of early-branching animals spanning from zygote formation (Z) to the blastula stage (B). Credits: Silhouettes were obtained from PhyloPic (https://www.phylopic.org/). Schmidtea polychroa, created by by M. A. Grohme under a CC0 1.0 Universal Public Domain licence; Caenorhabditis elegans, created by B. Goldstein, vectorization by J. Warner under a CC0 1.0 Universal Public Domain licence; Isohypsibius dastychi, created by B. Lang under a CC0 1.0 Universal Public Domain licence; Sophophora melanogaster, created by A. Wilson under a CC0 1.0 Universal Public Domain licence; Strongylocentrotus purpuratus, created by C. Schomburg under a CC0 1.0 Universal Public Domain licence; Danio rerio, created by Ian Quigley under a CC BY 3.0 licence; Nematostella vectensis, created by J. Warner under a CC0 1.0 Universal Public Domain licence; Amphimedon queenslandica obtained under a CC0 1.0 Universal Public Domain licence; Mnemiopsis leidyi, created by J. R. Winnikoff under a CC0 1.0 Universal Public Domain licence; Creolimax, created by Y. Wong using scanning electron microscopy images by A. Sebé-Pedrós (public domain agreed by I. Ruiz-Trillo) under a licence free of copyright, CC PDM 1.0.
Extended Data Fig. 3 Symmetry breaking during early development of C. perkinsii.
(a) Cleavage division dynamics at constant volume over time, visualized using a plasma membrane (PM) marker FM4-64 (Supplementary Video 4). Scale bar, 10 µm. (b) PM-stained live late colonies exhibiting internal cavities (arrows). This result has been reproduced at least 2 independent times. Scale bar, 10 µm. (c) U-ExM stained cells for microtubules (green), and DNA (blue), highlighting nuclear migration to the cortex prior to 1st mitotic division. This result has been reproduced at least 3 independent times. Scale bar, 10 µm. (d) Actin (magenta) and DNA-stained (blue) colonies at various cell stages, accompanied by volumetric segmentation of cells, highlighting the asymmetry in volume following the first cleavage division. This result has been reproduced at least 3 independent times. Scale bar,10 µm. (e) U-ExM stained cell for pan-labelling with NHS-Ester (red), microtubules (green), and DNA (blue), illustrating the first mitotic division at the cortex. This result has been reproduced at least 3 independent times. Scale bar, 10 µm. (f) Actin (magenta) and DNA-stained (blue) colonies at distinct cell stages showing the asymmetrical cell division in volume and time between the Aa and Ab cells. This result has been reproduced at least 3 independent times. Scale bar, 10 µm. (g) U-ExM stained colonies for pan-labelling with NHS-Ester (red), microtubules (green), and DNA (blue), highlighting a three-cell stage. This result has been reproduced at least 3 independent times. Scale bar, 10 µm.
Extended Data Fig. 4 Distinct cell fates following the release of C. perkinsii’s colonies.
(a) Time-lapse images over 14 h of new born C. perkinsii cells post-release show a cell-size increase in proliferative/mitotic cells (black arrows) and no increase in flagellated cells (white arrows) (Supplementary Video 7). Scale bar: 10 µm. (b) A short-duration high-resolution time-lapse of new born C. perkinsii cells post-release (T = 104 h) shows a cell-size increase in proliferative/mitotic cells (black arrows) and no increase in flagellated cells (white arrows) (Supplementary Video 8). Bar, 10 µm. (c) Kymographs of both flagellated (F) and proliferative/mitotic (P/M) cells over 10 min show the increase in cell size for the P/M cell. Bar, 10 µm. (d) Cell area over time of single cell traces reveals the increase in size for the proliferative/mitotic (P/M) cells in contrast to the flagellated cells (F) (neach = 5 cells). (e) Growth ratio between t = 0 and t = 10 min for both proliferative/mitotic (P/M) and flagellated cells (F) (nF = 5, nP/M = 5 cells). Statistical analysis using a two-tailed Student’s t-test, **P = 0.001. Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value. (f) U-ExM stained cells for pan-labelling with NHS-Ester (red), microtubules (green), and DNA (blue), identifying proliferative/mitotic and flagellated cells, as well as cells undergoing mitosis. Scale bar: 10 µm. Zoomed-in insets represent Z-stacks of each cell. Scale bar: 1 µm. (g) Nuclear volume measured from U-ExM images for both proliferative/mitotic (P/M) and flagellated cells (F) (nF = 10, nP/M = 16 cells). Statistical analysis using a two-tailed Student’s t-test, **P = 0.00016. Box plots extend from the 25th to 75th percentile, including the median, and whiskers extend from the minimum to maximum value. (h) Image analysis workflow and quantifications showing the spatial clustering of flagellated cells within multicellular colonies (see methods). Two maximum projections per cell (upper and bottom halves) were split into four quarters (Q1, Q2, Q3, Q4). Flagellated cells were counted in each quarter, and the quarter with the highest number was aligned to Q1. Mean and standard deviation were calculated and represented (neach = 5 cells).
Supplementary information
Supplementary Video 1
Time-lapse of two synchronized cells of C. perkinsii obtained with epifluorescent microscopy. The time interval between frames is 1 h. The video is played at 7 fps. Both cells can be seen undergoing a full life cycle with the release of new-born cells. Scale bar, 15 µm.
Supplementary Video 2
Time-lapse of distinct cell types: amoeboflagellate (left) harbouring a flagellum and exhibiting cellular protrusions and a mitotic cell without a flagellum (right) undergoing seemingly cell division. The time interval between frames is 5 s. The video is played at 7 fps. Scale bar, 5 µm.
Supplementary Video 3
Flagellated cells freely moving around following cell release among other non-flagellated cells. The time interval between frames is 1 s. The video is played at 7 fps. Scale bar, 10 µm.
Supplementary Video 4
Time-lapse of a Plasma Membrane-stained (PM) C. perkinsii cell undergoing a series of cleavage divisions at constant volume and showcasing cortical rotations. The time interval between frames is 15 min. The video is played at 7 fps. Scale bar, 10 µm.
Supplementary Video 5
Z-stacks of PM-stained live colonies at distinct cell stages, highlighting the patterned cleavage divisions, tetrahedral four-cell stage and formation of spatially organized multicellular colonies. Scale bar, 10 µm.
Supplementary Video 6
Z-stacks of two U-ExM stained colonies for pan-labelling with NHS-Ester (red), microtubules (green) and DNA (blue) at the three-cell stage. Scale bar, 10 µm.
Supplementary Video 7
Time-lapse video of new-born C. perkinsii cells post-release obtained with epifluorescent microscopy. The time interval between frames is 1 h. The video is played at 7 fps. We can observe the proliferating/mitotic cells rapidly increasing their cell size, whereas flagellated cells do not, following release. Scale bar, 10 µm.
Supplementary Video 8
Time-lapse video of new-born C. perkinsii cells post-release obtained with epifluorescent microscopy with a time interval of 1 s. The video is played at 7 fps. Proliferative/mitotic cells rapidly increase in size, whereas flagellated cells do not. Scale bar, 10 µm.
Supplementary Video 9
Three-dimensional visualization of U-ExM stained late colonies for pan-labelling with NHS-Ester (grey), microtubules (red) and DNA (cyan), highlighting the co-existence and clustering of flagellated and non-flagellated cells in the multicellular colony. The bar is uncorrected for an expansion factor of 4.2×.
Supplementary Video 10
Thhree-dimensional visualization of U-ExM stained late colonies for pan-labelling with NHS-Ester (grey), microtubules (red) and DNA (cyan), highlighting the co-existence and clustering of flagellated and non-flagellated cells in the multicellular colony. The bar is uncorrected for an expansion factor of 4.2×.
Supplementary Table 1
Gene ontology term enrichment for the five different gene expression clusters throughout the life cycle of C. perkinsii.
Supplementary Table 2
C. perkinsii genes associated with enriched gene ontology terms (one-sided Fisher’s exact test, topGO weight algorithm) for the five different gene expression clusters plus enriched gene ontology terms (one-sided Fisher’s exact test, topGO weight algorithm) of highly correlated genes among early-diverging animals (distribution of correlations shown in Extended Data Fig. 2g) and all animals (distribution of correlations shown in Extended Data Fig. 2h).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Olivetta, M., Bhickta, C., Chiaruttini, N. et al. A multicellular developmental program in a close animal relative. Nature 635, 382–389 (2024). https://doi.org/10.1038/s41586-024-08115-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08115-3
This article is cited by
-
A smartphone analogy to explore the origin of animals
The EMBO Journal (2026)
-
The evolutionary origins of synaptic proteins and their changing roles in different organisms across evolution
Nature Reviews Neuroscience (2026)
-
The evolutionary foundations of transcriptional regulation in animals
Nature Reviews Genetics (2025)
-
How did life get multicellular? Five simple organisms could have the answer
Nature (2025)
-
Morphogenesis of Fractofusus andersoni and the nature of early animal development
Nature Communications (2025)