Abstract
Substantial epigenetic resetting during early embryo development from fertilization to blastocyst formation ensures zygotic genome activation and leads to progressive cellular heterogeneities1,2,3. Mapping single-cell epigenomic profiles of core histone modifications that cover each individual cell is a fundamental goal in developmental biology. Here we develop target chromatin indexing and tagmentation (TACIT), a method that enabled genome-coverage single-cell profiling of seven histone modifications across mouse early embryos. We integrated these single-cell histone modifications with single-cell RNA sequencing data to chart a single-cell resolution epigenetic landscape. Multimodal chromatin-state annotations showed that the onset of zygotic genome activation at the early two-cell stage already primes heterogeneities in totipotency. We used machine learning to identify totipotency gene regulatory networks, including stage-specific transposable elements and putative transcription factors. CRISPR activation of a combination of these identified transcription factors induced totipotency activation in mouse embryonic stem cells. Together with single-cell co-profiles of multiple histone modifications, we developed a model that predicts the earliest cell branching towards the inner cell mass and the trophectoderm in latent multimodal space and identifies regulatory elements and previously unknown lineage-specifying transcription factors. Our work provides insights into single-cell epigenetic reprogramming, multimodal regulation of cellular lineages and cell-fate priming during mouse pre-implantation development.
Similar content being viewed by others
Main
After fertilization, cells undergo considerable epigenetic reprogramming for zygotic genome activation (ZGA) and lineage specification to generate transient totipotent cells and the lineage tree towards the inner cell mass (ICM) or the trophectoderm (TE)4. In mice, totipotency is limited to cells in the zygote and two-cell stages5. Meanwhile, studies have shown that the two cells that result from the first cleavage division exhibit cellular heterogeneities and uneven developmental potential6,7. Low-input profiling of histone modifications has also revealed a dynamic epigenomic landscape during early mammalian embryo development. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) presents non-canonical broad distribution until the late two-cell stage8,9,10, whereas H3K27me3 is depleted from promoter regions before blastocyst formation9,11. H3K9me3 undergoes large-scale re-establishment after fertilization, and the imbalance between two parental genomes lasts until the blastocyst stage12,13,14,15. Various single-cell multiomic methods for analysing chromatin accessibility, DNA methylation and gene expression have been used to interrogate allelic-specific reprogramming of multilayered epigenetic information16,17. However, despite improvements in the low-input profiling of histone modifications and chromatin accessibility18, a single-cell genome-coverage landscape of core histone modifications that influence the lineage tree is yet to be achieved. Completion of this task would provide important information for our understanding of epigenetic mechanisms that underlie the link between cellular heterogeneities and lineage specification.
TACIT has high genome coverage
To elucidate in detail how core histone modifications affect cellular heterogeneity, we established TACIT for single-cell profiling of the epigenome. TACIT is based on our in situ chromatin immunoprecipitation with sequencing (ChIP–seq) method19, and it produced similar numbers of reads per cell to that of bulk measurement (Fig. 1a). Notably, TACIT generated a 41-fold increase in non-duplicated reads per cell (Extended Data Fig. 1a–c). We first conducted TACIT experiments in mouse embryonic stem (ES) cells to profile H3K4me3, acetylation on histone H3 lysine 27 (H3K27ac), H3K36me3 and H3K27me3 modifications (Supplementary Tables 1 and 2). The aggregate profiles for each modification closely mirrored those generated by bulk ChIP–seq, but with high signal-to-noise ratios (Extended Data Fig. 1d,e), as demonstrated by the high fraction of reads in peaks (Extended Data Fig. 1f). TACIT also generated more non-duplicated reads than other single-cell methods that profile histone modifications19,20,21,22,23 (Extended Data Fig. 1g). We also noted the considerable proportion of TACIT reads mapped to mitochondrial DNA, a result that is primarily due to its higher sequencing depth than other single-cell profiling methods of histone modifications (Supplementary Table 3).
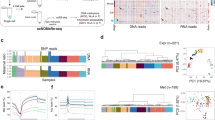
a, Schematic of the TACIT workflow. Cells were lightly fixed in cold methanol to retain intact nuclei. Permeabilized cells were incubated with antibodies, PAT–MEA/B (PAT assembled with MEA and MEB adaptors) and tagmentation buffer before manual pipetting of single cells into a well of 96-well plate. A brief lysis step (at 55 °C for 15 min with 0.1 mg ml–1 proteinase K) was key to minimize loss of material and to obtain genome-coverage reads per cell. b, Schematic of the TACIT experimental design in early mouse embryos. Cells from zygote, 2cell, 4cell, 8cell, morula and blastocyst stages were collected and subjected to genome-wide localization profiling of histone modifications as indicated at single-cell resolution with TACIT. c, Track view showing TACIT signals of various histone modifications in mouse embryos. Public datasets for H3K4me3, H3K27ac, H3K36me3, H3K27me3, H3K9me3 and H2A.Z were downloaded from the NCBI Gene Expression Omnibus (GEO) database (accessions GSE71434, GSE72784, GSE207222 (both H3K27ac), GSE112835, GSE76687, GSE97778 and GSE51579, respectively). Agg, aggregate. d, Violin plots displaying the distribution of non-duplicated reads per cell for each histone modification across different stages. The median number of non-duplicated reads for each stage from at least three independent experiments are shown on the top. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). e, UMAP visualization of high-quality single-cell data of H3K4me1 (n = 392), H3K4me3 (n = 635), H3K27ac (n = 538), H3K27me3 (n = 549), H3K36me3 (n = 579), H3K9me3 (n = 496) and H2A.Z (n = 560) modifications. Each dot represents an individual cell and is coloured by stages (left) and clusters (C1–C4; right). f, Euclidean distance between individual cells for each histone modification across different stages.
TACIT across early embryo development
We applied TACIT to generate genome-wide maps of histone modifications for embryos from the zygote, two-cell (2cell), four-cell (4cell), eight-cell (8cell), morula and blastocyst stages (Fig. 1b and Extended Data Fig. 2a,b). We ensured that the number of cells profiled for each stage was 5–50 times the actual number of embryonic cells at the respective stage. To fully cover potential regulatory elements and genic regions, we measured seven histone modifications that are known to be located at promoters (H3K4me3), enhancers (H3K4me1 and H3K27ac), gene bodies (H3K36me3) and heterochromatin (H3K27me3 and H3K9me3), as well as a histone variant (H2A.Z), in cultured cells24 (Fig. 1c). In total, we collected TACIT data from 3,749 cells: 392 for H3K4me1, 635 for H3K4me3, 538 for H3K27ac, 549 for H3K27me3, 579 for H3K36me3, 496 for H3K9me3 and 560 for H2A.Z (Supplementary Tables 2–4). Overall, we obtained up to half a million non-duplicated reads per cell for H3K4me1 at the 2cell stage (Fig. 1d). Of note, TACIT can be implemented with as few as 20 cells.
High correlation was obtained across different TACIT experiments (Extended Data Fig. 2c). Similarly, the aggregated TACIT profiles closely resembled available low-input bulk ChIP–seq datasets for H3K4me3, H3K9me3 and H3K27ac (Extended Data Fig. 2d). There was a clear decrease in the median number of non-duplicated reads per cell in zygotes compared to blastocysts for active marks such as H3K4me3 (233,164 versus 23,272), H3K4me1 (261,716 versus 129,603), H3K27ac (98,559 versus 53,563) and H3K36me3 (100,594 versus 49,146) (Fig. 1d). This observation was consistent with the shift from a broad to a sharp distribution in aggregate track views (Extended Data Fig. 2e). We also used indexing and tagmentation-based ChIP–seq (itChIP–seq)20 to independently confirm the quality of the data (Extended Data Fig. 2f,g). Overall, we obtained high coverage of the entire genome in single-cell profiles of histone modifications in the early embryo (Extended Data Fig. 2h,i).
Next, we performed clustering across each stage (Fig. 1e). Cells were predominantly clustered by histone modifications on the basis of developmental stages, a result that reflects the extensive epigenetic reprogramming that occurs during development. However, we also found varied heterogeneity among stages for each histone modification (Extended Data Fig. 3a,b). To further assess the degree of heterogeneity at each stage, we calculated the median Euclidean distance between cells, which was normalized against that observed among zygotes. There was a gradual increase in variation in cells from the zygote to 4cell stages (Fig. 1f). Notably, H3K27ac profiles exhibited marked heterogeneity as early as in single cells of the 2cell stage, with the following scaled median distance for each stage: 1 (zygote), 6.77 (2cell), 6.53 (4cell), 6.75 (8cell), 8.20 (morula) and 7.26 (blastocyst). This finding was in contrast to other profiles such as H3K4me3 (1 (zygote), 0.70 (2cell), 5.10 (4cell), 4.88 (8cell), 6.83 (morula) and 4.85 (blastocyst)), H3K36me3 (1 (zygote), 2.66 (2cell), 10.09 (4cell), 3.12 (8cell), 4.93 (morula) and 2.40 (blastocyst)) and H3K4me1 (1 (zygote), 1.13 (2cell), 2.02 (4cell), 0.99 (8cell), 1.57 (morula) and 1.26 (blastocyst)), which did not show substantial heterogeneity until the 4cell stage. This observation suggests that cells in the 2cell stage may start to display heterogeneity by establishing H3K27ac.
TACIT and CoTACIT in synthetic single cells
The use of combined histone modification profiles to annotate chromatin states has emerged as a powerful method for discovering regulatory elements without previous knowledge25,26,27. To investigate the dynamics of chromatin states during development, we integrated profiles of the six histone modifications analysed here into the same single cell on the basis of the correlation between histone modifications and gene expression. H2A.Z profiles were excluded from downstream analyses owing to ambiguous gene regulation with transcription28,29. Because of the inverse correlation observed between repressive histone modifications (H3K27me3 and H3K9me3) and gene expression, we developed a combined assay of target chromatin indexing and tagmentation (CoTACIT) to simultaneously profile multiple histone modifications in the same single cell. CoTACIT involved several rounds of antibody binding, protein A-Tn5 transposon (PAT) incubation and tagmentation to simultaneously measure multiple histone modifications (Fig. 2a). We generated H3K27ac–H3K27me3–H3K9me3 CoTACIT profiles from a total of 659 cells spanning 6 stages (Extended Data Fig. 4a and Supplementary Tables 2–4). Aggregated profiles between TACIT and CoTACIT showed good agreement for each histone modification (Extended Data Fig. 4b,c). Co-embedding of the TACIT and CoTACIT results also confirmed the quality of the data (Fig. 2b). Taken together, these findings indicate that CoTACIT can measure multiple histone modifications in the same cell in an effective manner.
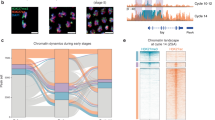
a, Schematic of the CoTACIT workflow. PAT-bar 1, PAT assembled with barcoded T5-1 and T7-1 adaptors; PAT-bar 2, PAT assembled with barcoded T5-2 and T7-2 adaptors. b, Co-embeddings of TACIT and CoTACIT data. Each dot represents an individual cell and is coloured by stages and methods. c, Schematic of the pipeline used to generate synthetic cells. d, Top, UMAP visualizations of interpolated 155 single cells based on WNN analysis, which combined all six modalities. Cells are coloured by stages. Bottom, boxplots displaying the average expression of ZGA-related genes for synthetic cells up to the 4cell stage. Genes exhibiting significant upregulation subsequent to the initiation of both minor and major ZGA were categorized as ZGA-related. e, Emission probabilities for each synthetic cell by single-cell ChromHMM. Chromatin-state definitions (left) and genome coverage (right) for each state are annotated. Chromatin-state definitions were determined on the basis of histone-modification probabilities and annotations of genic and non-genic elements (Extended Data Fig. 7a). f, Gene expression associated with chromatin states. Chromatin regions were linked to the nearest genes using Homer. The following number of genomic bins were used: multivalent (Multi), 278; promoter (weak) (Pr-W), 2,920; promoter (strong) (Pr-S), 36,352; enhancer (weak) (En-W), 16,026; enhancer (strong) (En-S), 59,573; gene body (poised) (Ge-P), 12,691; gene body (active) (Ge-A), 20,314; heterochromatin (polycomb) (He-P), 15,246; heterochromatin (H3K9me3) (He-K9), 11,913; and heterochromatin (K27+K9) (He), 23,039. g, Track view displaying chromatin-state annotations in representative loci for synthetic cells. Colours are as for e. For boxplots (d,f), the centre lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× the interquartile range (IQR).
We developed a workflow that included RNA-anchored multimodality integration to achieve six histone modification profiles in single cells (Fig. 2c). First, we generated single-cell RNA sequencing (scRNA-seq) data from 1,012 cells across early development, which produced a median of 9,583 genes identified per cell (Extended Data Fig. 5a and Supplementary Tables 2 and 4). On the basis of the overall positive correlation between active histone modifications and transcription30 (Extended Data Fig. 5b), we applied the workflow in Seurat31 to integrate RNA profiles with H3K4me1, H3K4me3, H3K27ac and H3K36me3 single-cell TACIT data and H3K27ac single-cell CoTACIT data separately (Extended Data Fig. 5c). Different histone modification profiles were therefore interpolated into the same RNA profile for each cell. Interpolated cell profiles revealed gradual chromatin remodelling during development (Fig. 2d), which was otherwise not evident by each modality individually (Extended Data Fig. 5d). Having acquired interpolated single-cell profiles, we performed clustering and defined synthetic single cells across developmental stages (Extended Data Fig. 5e,f). In brief, the similarity score among 155 interpolated cells was calculated to cluster cells on the basis of the multimodal epigenome. This resulted in 1, 2, 4, 8, 16 and 59 synthetic cells at the stages of zygote, 2cell, 4cell, 8cell, morula and blastocyte embryos, respectively. Overall, we observed mutually exclusive localization of active and repressive histone modifications in synthetic cells on selected genomic regions (Extended Data Fig. 5g–j). We found higher expression in ZGA-related genes32 in 2cell 2 than in 2cell 1 synthetic cells (Fig. 2d).
We used ChromHMM33 for each synthetic cell to annotate chromatin along the developmental trajectory. We observed an increased proportion of reads mapped to mitochondrial DNA in embryo profiles (Supplementary Table 3), a result that reflects the high copy number of mitochondrial DNA present in pre-implantation embryos34. All reads from mitochondrial DNA were filtered out in subsequent analyses. We segmented the genome and trained a 12-state model for each synthetic cell (Fig. 2e). Each state was assigned a descriptive label on the basis of its combination of histone modifications and overlap in the main genome categories (Extended Data Fig. 6a). These 12 chromatin states were placed into the following categories: multivalent (enriched for all histone modifications); promoters (enriched for H3K4me3 and H3K27ac); enhancers (enriched for H3K4me1 and H3K27ac); gene bodies (enriched for H3K36me3); and heterochromatin (enriched for H3K27me3 or H3K9me3). Notably, when synthetic cells were generated through a random shuffle of interpolated cell profiles, the chromatin states were comparable between the two cells analysed (Extended Data Fig. 6b), which confirmed the validity of intra-stage cell heterogeneity.
We next explored the establishment of chromatin states during development. We identified a state, termed multivalent, that was present exclusively in cells before ZGA. We confirmed the presence of histone modifications in multivalent regions in TACIT single cells, which indicated that this result is not an artefact from the integrative analysis (Extended Data Fig. 6c). Further analysis revealed that 57% of multivalent regions were located in intergenic and intron regions (Extended Data Fig. 6d). We posited that these multivalent regions may prime the epigenome for gene activation. Indeed, more than half of these regions transitioned into active chromatin states in subsequent stages (Extended Data Fig. 6e). In support of this result, the acquisition of a multivalent state for zygotes was also observed in bulk low-input ChIP–seq data from both itChIP–seq and public datasets for ChromHMM (Extended Data Fig. 6f).
Genes near ChromHMM-defined promoters, enhancers or gene bodies exhibited significantly higher expression than around heterochromatin (Fig. 2f). The substantial remodelling of chromatin states during early development was consistent with transcriptional reprogramming, such as the induction of ZGA and establishment of pluripotency (Fig. 2g). We attempted to identify potential enrichment of transcription factors (TFs) in promoters, enhancers and gene bodies for each synthetic cell (Extended Data Fig. 6g and Supplementary Table 5). Because most of the ChromHMM-annotated regions were broad, we intersected these regions with peaks identified using assay for transposase-accessible chromatin using sequencing (ATAC-seq) and used Homer to call TF motifs. As expected, several known essential TFs, including NR5A2, ZSCAN4, ZSCAN29, DUX, SOX2, POU5F1, KLF4, ESRRB, EOMES, TEAD4, GATA6, GATA4, GATA3, CDX2 and PRDM15, were enriched around enhancers, promoters and gene bodies of corresponding stages. Moreover, many pluripotency-related TFs were transiently enriched in the 2cell 2 synthetic cell, presumably owing to genome-wide activation following ZGA.
Epigenetic heterogeneity at the 2cell stage
At the 2cell stage, heterogeneity of the two cells was observed in both monomodal (Fig. 1f) and multimodal analyses (Fig. 2d,e). In these experiments, 2cell embryos were collected 43–45 h after human chorionic gonadotropin (hCG) administration, and therefore consisted of late-stage 2cell embryos. As such, heterogeneity may be due to the two cells being in asynchronous developmental stages, even in the same group of early-stage, mid-stage and late-stage 2cell embryos. To minimize this influence, we profiled histone modifications at the early 2cell stage (30 h after hCG administration), as cells in this stage are not expected to initiate the major ZGA program35 (Extended Data Fig. 7a). We performed CoTACIT for joint profiles of H3K4me3, H3K27ac and H3K27me3 in 89 cells from the early 2cell stage. We achieved a relatively high number of non-duplicated reads per cell, although the number was lower than when TACIT was used to individually assay each mark (Extended Data Fig. 7b). CoTACIT and TACIT data displayed good agreement for each histone modification (Extended Data Fig. 7c,d). We also used Cramér’s V similarity method to quantify the degree of co-enrichment between different histone modifications in the same single cell. This analysis showed that there was low co-enrichment around different genomic regions (Cramér’s V similarity < 0.04) across all combinations (Extended Data Fig. 7e).
Next, we clustered cells on the basis of H3K27ac, H3K4me3, H3K27me3 or weighted nearest neighbour (WNN) integration of the three modalities. Two major clusters (2cell 1 and 2cell 2) were successfully distinguished and corresponded to low or high ZGA scores (Fig. 3a,b). Notably, uniform manifold approximation and projection (UMAP) embedding of H3K27ac profiles from both TACIT and CoTACIT data showed similar heterogeneity in both early 2cell and late 2cell states (Extended Data Fig. 7f). This result provides support for the idea that the onset of heterogeneity in chromatin states occurs as early as the 2cell stage. This cellular heterogeneity at the early 2cell stage was not detected by transcription alone (Extended Data Fig. 7g). CoTACIT data showed that the 2cell 1 cell population had a significantly higher breadth score than the 2cell 2 cell population for both H3K4me3 and H3K27ac marks (Fig. 3c). We defined a breadth score by calculating the fraction of reads in the broad domain in zygotes for each single cell (Extended Data Fig. 7h). These results confirmed the broad-to-sharp transition of H3K4me3 and H3K27ac at the late 2cell stage, which were in line with results from the TACIT experiments (Extended Data Fig. 7i). Therefore, our results support the correlation between ZGA and the broad distribution of histone modifications.
a, Connected UMAP visualization for early 2cell CoTACIT data (n = 89). Lines connect the same cells across modalities. Cells are coloured by WNN clusters. b, Violin plots showing histone-modification signals around major ZGA genes in 2cell 1 and 2cell 2 clusters. c, Track views of histone modifications on a representative locus in 2cell 1 (n = 55) and 2cell 2 (n = 34) single cells, with violin plots showing breadth scores across all genomic peaks. Box plots indicate the median (centre line), quartiles (limits) and 1.5× IQR (whiskers). NS, not significant. d, The workflow of in vivo and IVF embryo-barcoded TACIT experiments. e, Top, UMAP plots of single cells from in vivo early and late 2cell embryos. The fraction of embryos for which two cells are assigned to the same or different clusters are coloured in the bar graph. Bottom, violin plots showing the major ZGA score. Early 2cell (H3K27ac, 144; H3K4me3, 172); late 2cell (H3K27ac, 92; H3K4me3, 78). f, Linked dot plots of major ZGA scores for cells from the same in vivo embryo, with the black line linking the average scores. g, Top, UMAP plots of single cells of IVF early and late 2cell embryos. The fraction of embryos for which two cells are assigned to the same or different clusters are coloured in the bar graph. Bottom, violin plots showing the major ZGA score. Early 2cell (H3K27ac, 166; H3K4me3, 138); late 2cell (H3K27ac, 114; H3K4me3, 106). h, Linked dot plots of major ZGA scores for cells from the same IVF embryo, with the black line linking the average scores. P values were calculated using two-sided Wilcoxon tests (b,c,e–h).
To further distinguish intra-embryo and inter-embryo heterogeneity at the 2cell stage, we developed embryo-barcoded TACIT, in which each individual embryo was indexed to track each cell of origin in the same embryo (Fig. 3d). We collected in vivo and in vitro fertilization (IVF) mouse embryos for embryo-barcoded TACIT, aiming to further mitigate the influence of asynchronous fertilization timing. Among in vivo mouse embryos, for 32% of early 2cell embryos (23 out of 72), the two cells were allocated to different clusters on the basis of H3K27ac profiles. Similarly, in 37% of late 2cell embryos (17 out of 46), cells exhibited comparable clustering. In the context of H3K4me3 profiles, 31% of early 2cell embryos (27 out of 86) had their two cells classified into different clusters, and this pattern of intra-embryo heterogeneity was also observed in 36% of late 2cell embryos (14 out of 39) (Fig. 3e). To further minimize such effects between embryos, we examined H3K27ac and H3K4me3 signals around ZGA genes for the two cells from the same embryos; again, we discovered a significant difference (Fig. 3f). These results were also observed in early and late 2cell IVF embryos (Fig. 3g,h). Collectively, these findings substantiate the presence of intra-embryo heterogeneity across both early and late 2cell developmental stages.
Earliest ZGA regulation revealed by CoTACIT
Given the detection of cell heterogeneity as soon as the early 2cell stage, we investigated the regulatory mechanisms that underpin the earliest stages of ZGA initiation. We first examined the dynamics of H3K4me3, H3K27ac and H3K27me3 marks around 720 ZGA-related gene regulators along the pseudotime (Extended Data Fig. 7j). To interrogate key regulatory elements involved in the earliest initiation of ZGA, we developed an analytical framework to link enhancers to promoters on the basis of the occurrence of both H3K4me3 in promoters (reads within ±5 kb flanking the transcription start sites (TSSs) of target genes) and H3K27ac in enhancers (reads in the distal regions) across cells (Extended Data Fig. 7k). Among the 43,983 putative promoter–enhancer pairs, promoters were typically regulated by multiple enhancers, with a median of six enhancers per promoter (Extended Data Fig. 7l). However, each enhancer was linked to a median of two promoters. These findings are consistent with the known complex multienhancer interactions in gene regulation36. To investigate promoter–enhancer pairs implicated in ZGA, we chose pairs for which H3K4me3 peaks fell within ±2 kb of the TSSs of genes that were activated after ZGA. We further divided the 1,812 ZGA-related promoter–enhancer pairs into three distinct groups: 2cell 1-specific pairs, 2cell 2-specific pairs and shared pairs (Extended Data Fig. 7m and Supplementary Table 6). The 2cell 1-specific pairs were associated with maternal genes such as Obox6. By contrast, 2cell 2-specific promoter–enhancer pairs were predominantly linked to ZGA-related genes, including Dppa4, Dppa3, Dppa5a and Zscan5b.
Murine endogenous retrovirus with leucine tRNA primer (MERVL) elements are known to be active in totipotency37. To examine what and how genes are regulated in the gain of totipotency, we chose promoter–enhancer pairs for which H3K4me3 or H3K27ac peaks fell in copies of MERVLs in which MERVLs may serve as putative promoters or enhancers, respectively (Extended Data Fig. 7n). MERVLs were defined as enhancers for more than 70% of the identified 1,188 MERVL-related promoter–enhancer interactions. Moreover, 51% of the identified MERVL-related promoter–enhancer interactions were activated after ZGA. Next, we identified TF motifs enriched in enhancers of the 322 promoter–enhancer pairs for which MERVL was defined as promoters (Extended Data Fig. 7o,p). In addition to the well-established totipotency TFs ZSCAN4 and DUX38,39, we found TFs such as NFYA, MEF2D, MAFK, SMAD3–SMAD5, POU5F1, POU2F3 and OBOX5. We also tested the possibility that MERVL may also function as enhancers to target non-MERVL genes (Extended Data Fig. 7o). Gene ontology (GO) term analysis showed that promoters regulated by MERVL were associated with DNA double-strand break repair (Extended Data Fig. 7q), which was also found in H3K9me3-marked transposable elements such as SINE-VNTR-Alu13.
Totipotency definition and driver TFs
We aimed to use integrated chromatin states to define the transient totipotent state in 2cell and 4cell stages. To this end, we adopted a classification approach and divided chromatin states into five groups: enhancers, promoters, gene bodies, polycomb-protein-associated heterochromatin and H3K9me3-associated heterochromatin. Using the posterior probability matrix, we performed dimensionality reduction with latent semantic indexing and UMAP, and clustered cells on the basis of each chromatin state separately. Our analysis revealed that chromatin states positively associated with transcription (promoters, enhancers and gene bodies) successfully resolved embryo stages regardless of whether all bins were used (Fig. 4a) or only those overlapping with TSSs (Extended Data Fig. 8a). Notably, both polycomb-protein-associated and H3K9me3-associated heterochromatin also exhibited the ability to separate cells on the basis of totipotent states.
a, UMAP plots of synthetic cells based on the posterior probabilities of ChromHMM-defined chromatin states. b, Schematic of the two strategies used for discovering potential classifier bins to define totipotency. Strategy 2 prioritizes 2,583 potential totipotency-related classifier bins. c, Venn diagram showing the overlap of totipotency-related classifier bins generated from the two machine-learning models. The P value was calculated using one-sided hypergeometric tests. d, Heatmaps displaying chromatin-state annotations in all 2,583 classifier bins for synthetic cells from strategy 2. The 2,583 classifier bins were grouped into two clusters using k-means clustering: one annotated as active in 2cell synthetic cells and the other as repressive in 8cell synthetic cells. e, TF motifs enriched on the active 2,583 classifier bins for each synthetic cell. The evaluation of totipotency by classifier bins for each synthetic cell is plotted on the right. TF motifs with both high enrichment (–log10(P) > 8) and expression (transcripts per million (TPM) > 2) in totipotent cells were selected as putative totipotency-related TFs and highlighted in bold. Representative, previously reported pluripotency-associated and lineage-associated TFs are also shown as control. P values were calculated using one-sided binomial tests. f, Ranking plots for the enrichment of different transposable elements in the 2,583 classifier bins. Enrichment was calculated using observed versus expected probability. Bold labels highlight key transposable elements that have been previously reported to be associated with totipotency. g, Heatmap showing the percentage of transposable-element copies that were annotated as promoter, enhancer, gene body, heterochromatin or quiescent/low states. Transposable elements enriched in the 2,583 classifier bins (log2(overexpression) > 1) are shown.
We focused on identifying genomic intervals that showed significant differences in chromatin annotation between cells at the 2cell and 8cell stages. The annotation of these differential genomic bins may help better define totipotency. Totipotency-defined differential bins annotated as promoters, enhancers and gene bodies were reduced during development, which was in good agreement with the expression dynamics of totipotency marker genes (Extended Data Fig. 8b). Conversely, differential bins annotated as heterochromatin states showed a marked increase, which highlighted an inverse correlation with totipotency.
Next, we developed a framework to identify feature classifier bins between totipotency-high and totipotency-low cells (Fig. 4b). We designed two strategies for identifying classifier bins between 2cell 1 with 2cell 2 synthetic cells (strategy 1) and 2cell with 8cell synthetic cells (strategy 2), in which the former strategy aimed to rule out any differences in stage. For each chromatin state, we selected genomic regions for which posterior probability values had correlations of greater than 80% or less than −50% with the expression of totipotency marker genes and integrated all highly correlated genomic regions. Next, the generated state matrix of cells was input into supervised machine-learning classifier tools40 to output relevant regions for defining totipotency. We identified 2,927 genomic regions with the potential to define totipotency with strategy 1 and 2,583 genomic regions with strategy 2, with 28.18% overlap between the two sets of classifier bins (Fig. 4c and Supplementary Table 7).
Chromatin-state annotations on these classifier bins already showed distinction for cells at the 2cell and 8cell stages (Fig. 4d). Overall, the 2,583 classifier bins from strategy 2 were highly enriched with transposable elements (Extended Data Fig. 8c). Moreover, 31% of the 1,338 classifier bins annotated as gene bodies at the 2cell stage also overlapped with known feature totipotency genes (Extended Data Fig. 8d). To explore TFs that potentially function during the gain or loss of totipotency, we identified TF motifs enriched in the active classifier bins for each synthetic cell (Fig. 4e and Supplementary Table 8). We used all 2cell and 8cell ATAC-seq peaks as background when assessing TF motif enrichment by Homer to disentangle the influence of open chromatin41. ZGA-associated TFs such as ZSCAN4 and NR5A2 were enriched in cells at the 2cell stage, whereas pluripotency-associated TFs such as ESRRB, KLF4, NANOG and POU5F1 were enriched in cells at the 8cell stage. After stringent filtering (Methods), we obtained 120 potential totipotency-related TFs. GO term analysis indicated that these TFs were related to the embryo development process (Extended Data Fig. 8e). Seven TFs (MEF2D, ALX1, CEBPG, LBX1, ETS2, ESR2 and ESR1) were enriched in cells annotated with high totipotency. Notably, the motif enrichment of these TFs was also positively correlated with gene expression (Fig. 4e). The TF motif enrichment of MEF2D, LBX1, ESR1, ETS2 and CEBPG was also observed in the 2,927 classifier bins from strategy 1 (Extended Data Fig. 8f), a result that supports the robustness of the method.
To verify whether candidate TFs identified in classifier bins are implicated in totipotency induction, we performed gain-of-function assays through CRISPR-mediated transcriptional activation (CRISPRa) targeting the promoter of the top seven enriched TFs (MEF2D, CEBPG, ETS2, ESR1, ESR2, ALX1 and LBX1; Supplementary Table 9), with ZSCAN4 and DUX used as positive controls (Extended Data Fig. 8g). Mouse ES cells expressing dCas9–VPR were infected with lentiviral vectors expressing single guide RNAS (sgRNAs), which were read out by scRNA-seq. Overall, about 48% of single cells contained 1–5 sgRNAs and about 50% more than 10 sgRNAs (Extended Data Fig. 8h,i). Notably, a large proportion of mouse ES cells subjected to CRISPRa were clearly separated from control mouse ES cells42 (Extended Data Fig. 8j,k). They seemed to be activated in a permissive totipotent state, with an increase in expression of totipotency-feature genes (Extended Data Fig. 8l). We further classified cells into pluripotent (1,292 cells), intermediate (8,634 cells) and totipotent-like cells (252 cells) along pseudotime (Extended Data Fig. 8m,n). To evaluate the role of the tested TFs in inducing the totipotency program, we first ranked genes and used the pipeline MUSIC to calculate perturbation correlations among them using cells that received only one sgRNA. CEBPG, LBX and ESR1 had comparable perturbation effects with the positive control (ZSCAN4D and DUX) and showed higher perturbation scores than other TFs (Extended Data Fig. 8o,p). To further explore combination effects of induction of the totipotency gene program in CRISPRa experiments, we calculated totipotency scores for cells with various combinations of TF perturbation in which a candidate TF was removed one at a time. The highest totipotency scores and latest pseudotemporal stages were observed for TF combinations in which ETS2, ALX1, MEF2D or ESR2 were depleted, which indicated that these TFs are less potent in inducing totipotency (Extended Data Fig. 8q,r). Together, our results indicate that CEBPG, LBX and ESR1 have pivotal roles in the totipotent state.
A large number of transposable elements have been shown to initiate transcription during mouse pre-implantation development, contributing to the establishment of totipotency and pluripotency and to the activation of the embryonic genome43. In addition to the well-known MERVL elements, we found an increased frequency of long terminal repeat (LTR) elements in these 2,583 classifier bins (Fig. 4f). Thus, to refine functional transposable elements in totipotency, we examined chromatin states of all 75 enriched transposable elements (log2(observed versus expected ratio) > 1; Supplementary Table 10). Overall, 41 of these were highly active before the 4cell stage, with 10–30% copies of them annotated as promoters in zygotes and then transforming into gene bodies at the 2cell stage (Fig. 4g). Among these, B2 Mm1a, RLTR4 MM-int and RLTR4 Mm were extremely active, which implies that they have a role in establishing totipotency.
Epigenetic priming of the first cell fate
During mouse pre-implantation development, the first lineage specification results in the formation of ICM and TE cells. Although this phase typically occurs during the blastocyst stage, recent studies have shown potential cell bias at earlier stages, implicating that there is cell fate pre-commitment44,45. We attempted to identify the earliest cell lineage priming for ICM and TE cells. To distinguish ICM and TE cells, we defined RNA cells as ICM or TE cells on the basis of marker-gene expression following clustering (Extended Data Fig. 9a). We transferred these labels to TACIT and CoTACIT cells after integrating RNA data with H3K4me1, H3K4me3, H3K27ac, H3K36me3 and CoTACIT H3K27ac modality. A comparison of these integrated multimodal profiles with public low-input bulk ChIP–seq data in blastocysts also resulted in good agreement (Extended Data Fig. 9b,c).
For chromatin-state annotation in single cells, we performed scChromHMM for integrated single cells46. We trained a 12-state HMM model based on aggregate ICM and TE profiles (Extended Data Fig. 9d) to annotate chromatin states for single cells. We merged five adjacent single cells along pseudotime into one synthetic cell before applying the forward–backward algorithm47. To build a lineage classifier and to predict the earliest lineage divergence defined by the epigenetic landscape, we established a random forest machine-learning model based on the chromatin-state annotation in ICM and TE synthetic cells (Fig. 5a), which produced a prioritization of 780 classifier bins (Supplementary Table 7). Notably, an area under the curve (AUC) value of 1.000 was achieved in the test group (Fig. 5b). All of the six histone modifications analysed seemed to be important for interrogating lineage specification, as depletion of any modality before training resulted in markedly decreased classification (Extended Data Fig. 9e). Overall, 37% of the 780 classifier bins were located around differentially expressed genes between ICM and TE cells (Fig. 5c), whereas the remaining 63% classifier bins may function as cis-regulatory elements for lineage specification. More than 80% of the 780 classifier bins were distant from TSSs (Extended Data Fig. 9f). GO term analysis indicated that these classifier bins were related to mouse embryonic development (Extended Data Fig. 9g).
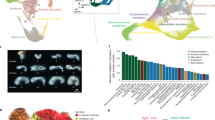
a, A computational pipeline for constructing a random forest training model to identify classifier bins associated with lineage specification. b, Receiver operating characteristic of the random forest model. c, Fraction of classifier bins (n = 780) overlapping with ICM and TE differential expressed genes (DEGs). d, Heatmap displaying chromatin-state annotations in all 780 classifier bins. The 780 classifier bins were grouped into four clusters using k-means clustering. e, TF motifs identified during ICM or TE lineage specification. TF motifs with high enrichment (–log10(P) > 5) and expression (reads per kilobase million (RPKM) > 1.5 in Ribo-lite data) along lineage specification are highlighted in bold (for TE) or underline (for ICM). P values were calculated using one-sided binomial tests. f, Quantification of early embryo development from 36 to 108 h. Sample sizes are as follows: control (37, 40, 24), NANOG (35, 22, 29), ZFX (25, 25, 33), HNF4A (38, 26, 24), YY2 (44, 25, 35), TCF12 (33, 34, 21), CEBPB (14, 37, 32), BBX (31, 20, 31), SMAD2 (43, 41, 42), HBP1 (34, 28, 36), CDX2 (34, 29, 41), KLF6 (31, 15, 35), SOX15 (41, 35, 15), MED1 (27, 17, 33), ELF5 (36, 36, 24), HIF1A (24, 14, 40). Data from three replicate experiments are shown for each time point. g, Quantification of morula embryos that develop into normal or abnormal blastocysts. Numbers inside each bar indicate the number of embryos. P values (shown on the chart) were calculated using two-sided Chi-square tests. h, Top, schematic of the two classes of abnormal blastocysts after KD. Bottom, quantification of abnormal blastocysts with SOX2– or CDX2– cell misallocation or the presence of ICM SOX2– cells. The total number of blastocysts is shown. P values were calculated using two-sided G-tests. i, Immunofluorescence staining of mouse embryos at 108 h after fertilization. Shown are z projection 3D images and single-section immunofluorescence images. Representative images out of three independent experiments are shown. Asterisks, adjacent embryos; white arrowheads, CDX2+SOX2+ cells; green arrowheads, misallocated CDX2+ cells. Scale bar, 100 μm.
We attempted to predict the lineage potential of cells at earlier stages. Using k-means clustering, we grouped the 780 classifier bins into TE-classifier and ICM-classifier bins (Fig. 5d). We identified distinct domains of active and inactive chromatin states, which together separated most cells with differentiation potentials into either the ICM lineage or the TE lineage. Notably, two 4cells, three 8cells and four morula cells bore more resemblance to ICM cells than to TE cells according to their chromatin-state annotations in the 780 classifier bins. This result suggests that these early epigenetic signatures may be pre-established in early stages before overt cell-fate bifurcation (Fig. 5d and Extended Data Fig. 9h). We next examined TF motifs enriched in ICM or TE classifier bins compared to open-chromatin regions along two lineage specification trajectories (Fig. 5e and Supplementary Table 8). In addition to previously reported functional TFs, such as CDX2, POU5F1, NANOG, PRDM15, GATA4 and FOXA2, we identified that HIF1A, ELF5, MED1, SOX15 and KLF6 were exclusively enriched at TE-potential morula and blastocyst cells, whereas HBP1, SMAD2, BBX, CEBPB, TCF12, YY2, HNF4A and ZFX were exclusively enriched at ICM-potential morula and blastocyst cells. This result suggests that these TFs may have roles in earliest cell-fate pre-determination. GO term analysis showed that these potential ICM-related or TE-related TFs were enriched for formation of primary germ layer and embryonic placenta development, respectively (Extended Data Fig. 9i).
To determine whether these identified TFs would be bona fide important regulators of ICM or TE cell-fate decision, we performed knockdown (KD) experiments against eight candidate TFs of ICM-cell fate and five TF candidates of TE-cell fate (a non-targeting short-interfering RNA (siRNA) was used as a control). RNA-seq confirmed that these TFs were knocked down by 34–92% at the 8cell and blastocyst stages (Extended Data Fig. 10a). KD of all 13 TFs reduced normal development towards blastocysts and resulted in fragmented or dead embryos (Fig. 5f and Extended Data Fig. 10b). We next assessed the rate of morula-to-blastocyst transition. KD of 12 TFs (BBX was not depleted) reduced the proportion of morula embryos developing into blastocysts (Fig. 5g). In addition, depletion of several potential TFs of ICM-cell fate (YY2, CEBPB and BBX) and TE-cell fate (KLF6, SOX15 and HIF1A) resulted in misallocation of SOX2+ or CDX2+ cells48 (Fig. 5h,i). Notably, Hnf4a and Smad2 KD led to the existence of SOX2– cells in ICM cells (Extended Data Fig. 10c). Further analysis of single-embryo RNA-seq data prioritized seven lineage regulators (YY2, CEBPB, SMAD2 and HNF4A for ICM-cell lineage, and SOX15, KLF6 and HIF1A for TE-cell lineage), as their loss of function significantly downregulated the expression of ICM and TE feature genes (Extended Data Fig. 10d–i). This result was verified through inspection of lineage-specific TF activity between the non-targeting control and KD of candidate TFs for ICM-cell or TE-cell fate (Extended Data Fig. 10j,k).
Discussion
In summary, we developed TACIT and CoTACIT to obtain genome-coverage single-cell histone modifications and joint multimodal profiles, respectively, for mouse development stages of zygotes to blastocysts. This epigenomic information-rich resource enabled us to identify cellular heterogeneities as soon as the early 2cell stage, to pinpoint chromatin states of key regulatory elements and potential TFs in defining totipotent cells and to backtrack cell-lineage potentials along the developmental path.
We found that a large fraction of prospective ICM and TE cells can be back tracked as early as the 8cell stage in our single-cell multimodal classification model. This finding is partially in line with results from studies that used live imaging for lineage tracing49,50. Furthermore, we identified eight TFs implicated in ICM-cell lineage specification and five TFs in TE-cell lineage specification, which were validated by in vivo loss-of-function experiments. We propose that our information-rich resource can be further integrated with other epigenomic modalities, such as DNA methylation and chromatin accessibility16,17, to gain more comprehensive insights into regulatory interactions in the epigenomic landscape.
Although our synthetic cell analysis revealed intra-embryo heterogeneity, a caveat that should be noted is that we sacrificed single-cell resolution in such an analytical pipeline. Further technology advancement in single-cell co-profiling of six or more modalities with similar genome-coverage may provide additional insights into epigenetic lineage regulation and tracing. Future studies are anticipated to elucidate epigenetic lineages and regulators in early human embryo development.
Methods
Animal use and care
All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Peking University. All mice were maintained in pathogen-free conditions at the Laboratory Animal Center of Peking University on a 12–12-h light–dark cycle, with a temperature of 20–25 °C and humidity of 30–70% and access to food and water ad libitum.
Single-cell isolation from mouse early embryos
To obtain pre-implantation embryos, superovulation was induced in 4-week-old C57BL/6J female mice through an intraperitoneal injection of 7.5 international units (IU) of PMSG (San-Sheng Pharmaceutical) followed by 7.5 IU of hCG (San-Sheng Pharmaceutical) 44–48 h later and then the mice were mated with 2-month-old C57BL/6J male mice. Each set of embryos at a specific stage was flushed from oviducts or uteri of pregnant female mice at the following defined time periods after hCG administration: 22–24 h (zygote), 30 h (early 2cell), 43–45 h (late 2cell), 54–56 h (4cell), 68–70 h (8cell), 78–80 h (morula) and 88–90 h (blastocysts). The embryos were maintained in M2 medium (Sigma). Germinal vesicle-stage oocytes were collected 48 h after PMSG administration.
To collect zygotes, a cumulus mass containing several zygotes surrounded by follicular cells was transferred to 1× hyaluronidase solution (Sigma) and incubated at 37 °C for a few minutes. The zygotes were then transferred to M2 medium and their zona pellucida was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. The second polar bodies of zygotes were manually removed with a very fine glass needle.
For embryos of other stages, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. The embryos were then transferred to a pre-warmed 1:3 mixture of TrypLE (Gibco) and Accutase (Gibco) and incubated at 37 °C for several minutes until the cell boundaries become apparent. The embryos were transferred to M2 medium and manually separated into single cells using a mouth pipette with an appropriate diameter needle. The dissociated embryonic cells were transferred to a pre-chilled 200 μl tube containing 10 μl cold 1% BSA–PBS and lightly fixed with 180 μl chilled methanol drop by drop. The cells were stored at −80 °C or immediately used for subsequent experiments. All tips and tubes used for cell collection were pre-rinsed with 0.1% BSA–PBS to avoid sample loss.
IVF embryo experiments
To collect embryos from IVF, oocytes were collected from C57BL/6J female mice 15 h after hCG injection. Oocytes were incubated in a 200 μl drop of HTF (M1135, Aibei) for 30 min before addition of the sperm suspension. Sperm samples were collected from C57BL/6J male mice and capacitated by placing in a 37 °C, 5% CO2 incubator for 60 min. Next, 3–5 μl of the sperm suspension taken from the edge of the sperm capacitation drop was added to the oocyte clutches (final sperm concentration of 1–5 × 105 cells per ml) and incubated for 3–4 h at 37 °C with 5% CO2. Forcefully pipetting the oocytes up and down several times in a 10 μl volume using a 200 μl pipette helped remove excess sperm. Viable fertilized oocytes were washed and transferred to a new 35 mm culture dish containing KSOM medium (M1435, Aibei). The embryos were distributed evenly throughout the culture dish and incubated at 37 °C with 5% CO2 overnight. The early 2cell and late 2cell embryos were collected at 20 and 35 h after IVF, respectively.
Mouse ES cell culture
Wild-type V6.5 mouse ES cells were cultured at 37 °C with 5% CO2 and were maintained on 0.1% gelatin-coated plates in high-glucose DMEM culture medium containing 15% fetal bovine serum (Invitrogen), 1% penicillin–streptomycin (Hyclone), 1% MEM nonessential amino acids (Cellgro), 1% Glutamax (Gibco), 1% nucleoside (Millipore), 0.1 mM 2-mercaptoethanol (Sigma) and 1,000 U ml–1 recombinant leukaemia inhibitory factor (Millipore).
Antibodies
The following antibodies were used for TACIT (catalogue and lot numbers provided after the supplier name): H3K4me1 (1:50; Abcam, ab8895, GR3369516-1); H3K4me3 (1:200; Millipore, 04-745, 3243412); H3K27ac (1:500; Diagenode, C15410196, A1723-0041D); H3K36me3 (1:200; Active Motif, 61101, 06221007); H3K27me3 (1:200; Millipore, 07-449, 3146226); H3K9me3 (1:200; Active Motif, 39161, 30220003); and H2A.Z (1:200, Abcam, ab4174, GR279096-1). Donkey anti-rabbit-Alexa 488 (1:500; Invitrogen, A32790) and donkey anti-rabbit-Alexa 555 (1:500, Invitrogen, A31572) were used as secondary antibodies. Antibodies used in immunofluorescence staining included SOX2 (1:200; Active Motif, 39843, 2226414) and CDX2 (1:200, BioGenex, MU392A-UC, MU392A0516D).
TACIT library generation and sequencing
TACIT produced more non-duplicated reads than other single-cell methods for profiling histone modifications (Extended Data Fig. 1g). This improvement was attributed to the following key modifications: (1) fixing cells with methanol rather than the widely used formaldehyde; (2) tagmenting cells with the high-activity PAT enzyme as experimentally titrated; (3) reducing loss of material by titrating the incubation time for reverse-crosslinking from hours to 15 min as well as rinsing tubes and plates with 0.1% BSA–PBS; and (4) performing a single-tube reaction after pipetting into a 96-well plate for better recovery. Specifically, methanol-fixed embryonic cells or mouse ES cells were placed on ice for at least 15 min for rehydration. Cells were washed twice with wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl and 0.5 mM spermidine (Sigma), 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) to remove residual methanol. We found that a brief centrifuge of cells at a low speed before aspirating the top two-thirds of the supernatant led to almost no cell loss during the washing procedure. This step ensured optimum cell recovery and satisfactory cell quality. In our experiment, different centrifugal speeds were applied to cells of different developmental stages because of the differences in the cell volume: 150g for zygotes, 200g for 2cell and 4cell stages, 350g for 8cell and morula stages and 1,000g for blastocysts.
Next, cells were incubated with specific antibody in 100 μl antibody buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine (Sigma), 2 mM EDTA, 0.01% digitonin, 0.05% TX-100, 1% BSA–PBS, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 3–4 h. After incubation, cells were washed twice with 180 μl Dig-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine (Sigma), 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) and suspended with 100 μl high-salt Dig-wash buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine (Sigma), 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) containing 3 μg ml–1 PAT–MEA/B. The PAT expression, purification and assembly procedures were performed as per previously described guidelines19. Cells were rotated at 4 °C for 1 h to enable complete binding of PAT to antibodies and then washed twice with 180 μl high-salt Dig-wash buffer to remove free PAT–MEA/B. Tagmentation was reactivated by suspending cells with 10 μl cold reaction buffer (10 mM TAPS-NaOH pH 8.3, 5 mM MgCl2, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) and incubated at 37 °C for 1 h in a PCR cycler. The reaction was stopped by adding 10 μl 40 mM EDTA and cells were washed twice with 1% BSA–PBS, and single cells were picked and placed into a well of a 96-well plate with a mouth pipette under a microscope. The 96-well plates were pre-rinsed with 1% BSA–PBS to avoid loss of DNA fragments, and 2 μl lysis buffer (10 mM Tris-HCl pH 8.5, 0.05% SDS and 0.1 mg ml–1 proteinase K) was added to each well. For each well, samples were covered with 5 μl mineral oil (Sigma) and incubated at 55 °C for 15 min to release DNA fragments. Next, 0.5 μl of 10 mM PMSF was added to each well to deactivate protease K, and 1 μl of 0.9% Triton X-100 was added to quench SDS in the reaction. Finally, 17 μl PCR mix (0.2 μl KAPA HiFi HotStart DNA polymerase, 4 μl 5× KAPA High-GC buffer, 0.5 μl 10 mM dNTP mix, 0.5 μl 25 mM MgCl2 and 10.8 μl H2O) was added to each well with 0.5 μl 10 mM Nextera i5 index primer and 0.5 μl 10 mM i7 index primer (Supplementary Table 1). PCR enrichment was performed in a thermal cycler with the following program: 1 cycle of 72 °C for 5 min; 1 cycle of 95 °C for 3 min; 11 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. The library was purified with 1× AMPure XP beads (Beckman) once, and 200–1,000 bp fragments were selected with 0.5× + 0.5× AMPure XP beads. The libraries were sequenced with paired-end 150-bp reads on a NovaSeq 6000 platform (Illumina).
CoTACIT library generation and sequencing
For CoTACIT with embryos, isolated single cells were rehydrated and washed as described above. For the first round of barcoding, cells were incubated with 0.5 μg H3K4me3 (for the early 2cell stage) or 0.5 μg H3K27ac (for all six developmental stages) in 100 μl antibody buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 2 mM EDTA, 0.01% digitonin, 0.05% TX-100, 1% BSA–PBS, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 3 h. Next, cells were washed twice with 180 μl Dig-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF). Cells were incubated with 3 μg ml–1 PAT-T5-1 and 3 μg ml–1 PAT-T7-1 in 100 μl high-salt Dig-wash buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine, 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 1 h and washed twice with 180 μl high-salt Dig-wash buffer. After tagmentation and inactivation with 20 mM EDTA, cells were washed 3 times with 180 μl 1% BSA–PBS to wash out free PAT and adapters. The second round of barcoding was performed as for the first round, except that cells were incubated with 0.5 μg H3K27ac (for the early 2cell stage) or 0.5 μg H3K27me3 (for all of six developmental stages) in 100 μl antibody buffer and barcoded with 3 μg ml–1 PAT-T5-2 and 3 μg ml–1 PAT-T7-2 in 100 μl high-salt Dig-wash buffer at 4 °C for 1 h. Similarly, the third round of barcoding was carried out using the same procedure, except that cells were incubated with 0.5 μg H3K27me3 (for the early 2cell stage) or 0.5 μg H3K9me3 (for all the six developmental stages) in 100 μl antibody buffer and barcoded with 3 μg ml–1 PAT-T5-3 and 3 μg ml–1 PAT-T7-3 in 100 μl high-salt Dig-wash buffer at 4 °C for 1 h. Finally, cells were washed 3 times with 1% BSA–PBS and single cells were picked and placed into a well of a prepared 96-well plate followed by fragment release, proteinase K inactivation and SDS quenching, as described for the TACIT procedure.
Two-round PCR was performed as previously described19, which resulted in the standard Illumina Truseq Compatible library. In brief, 20 μl PCR mix (0.2 μl KAPA HiFi HotStart DNA polymerase, 4 μl 5× KAPA high-GC buffer, 0.5 μl 10 mM dNTP Mix, 0.5 μl 25 mM MgCl2, 11.6 μl H2O and 0.5 μl 50 µM in total first-round primer mix) was added to each well. PCR was performed as follows: 1 cycle of 72 °C for 5 min; 1 cycle of 95 °C for 3 min; 8 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. Excess primers were digested by adding 0.25 μl ExoI (NEB) and plates were incubated at 37 °C for 60 min followed by 72 °C for 20 min. A volume of 10 μl second-round PCR mix (0.1 μl KAPA HiFi HotStart DNA polymerase, 2 μl 5× KAPA High-GC buffer, 0.25 μl 25 mM MgCl2 and 6.4 μl ddH2O) was added to each well containing 0.5 μl of 10 mM Truseq index i5 and 0.5 μl Truseq index i7 (Supplementary Table 1) and subjected to PCR with the following program: 1 cycle of 95 °C for 3 min; 5 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. The library was purified with 1× AMPure XP beads (Beckman) once and 200–1,000 bp fragments were selected with 0.5× + 0.5× AMPure XP beads. The libraries were sequenced with paired-end 150-bp reads on a NovaSeq 6000 platform (Illumina).
Embryo-barcoded TACIT
For embryo-barcoded TACIT with cells of early and late 2cell stages, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for 30 s. The embryos were transferred to M2 medium and directly fixed with methanol as described above. The embryos were stored at −20 °C or immediately used. Whole embryos were directly applied to the regular TACIT pipeline as described above. After tagmentation, the two cells from the same embryo were separated with custom microdissection needles and deposited into different wells of lysis buffer. Each well was covered with 5 μl mineral oil (Sigma) and incubated at 55 °C for 15 min to release DNA fragments. Next, 0.5 μl of 10 mM PMSF was added to each well to deactivate protease K, and 1 μl of 0.9% Triton X-100 was added to quench SDS in the reaction. PCR amplification was conducted as described for TACIT libraries, and DNA fragments of the two cells from the same 2cell embryos were barcoded with different combinations of Nextera i5 and i7 indexes. Finally, the embryo-barcoded TACIT library was sequenced as described for conventional TACIT.
Low-input itChIP
The itChIP–seq20 protocol was performed with a few modifications. First, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. Subsequently, embryos were transferred to M2 medium and fixed with 1% formaldehyde solution at room temperature for 3 min followed by 1× PBS wash and centrifugation at 4 °C. Samples were preserved at −80 °C or used immediately. Fixed embryos were incubated in hypotonic buffer (20 mM HEPES pH 7.9, 10 mM KCl, 10% glycerol, 0.2% NP-40 and 0.05% SDS) at 37 °C for 30 min to release chromatin. Embryos underwent gentle fragmentation by sonication (Q800R sonicator, 20% power, 10 s) and quenched with Triton X-100. Genomic tagmentation was obtained by incubating with Tn5 assembled with MEA/B adapters at 37 °C for 1 h. After the tagmentation reaction, samples were further processed to release chromatin from nuclei. After centrifugation at 4 °C, the soluble supernatant was isolated and incubated with antibodies overnight. Dynabeads protein A (Invitrogen) beads were used to pull down chromatin–antibody complexes. DNA fragments were eluted from beads and treated with proteinase K. The resultant DNA was purified and extracted using phenol–chloroform, followed by library preparation using the KAPA HiFi HotStart technique as per the TACIT procedure and supplemented with Illumina Nextra index primers. After size selection for fragments ranging from 200 to 1,000 bp, the libraries were quantified using Qubit to determine their concentration. The pooled samples were sequenced on a NovaSeq 6000 (Illumina) for paired-end 150 bp reads.
Microinjection in zygotes for siRNA knockdown
For siRNA knockdown, isolated zygotes were microinjected with sets of three siRNAs against targets (20 μM in total) or with non-target control (NC, 20 μM in total). The following siRNAs were used: NC, UGGGACUUGCAGGCCUGAUAUTT; Nanog, CGAGAACUAUUCUUGCUUATT, CCUGAGCUAUAAGCAGGUUAATT and UGGAGUAUCCCAGCAUCCAUUTT; Zfx, GGUUCAUGAUAGUGUAGUATT, GGAUGAAGAUGGACUUGAATT and GGAGGACAACGAAAUGAAATT; Yy2, GCUGCGAGAAGAUGUUCAATT, CACCAUGUGGGACGAUGUUAATT and GACCUAUAGCAUGCUCUCAUATT; Tcf12, GUGGCAGUCAUCCUUAGUCUATT, GAUGCAAUGUCCUUCUUAATT and GGAACAAGUGGUCAACCAATT; Cebpb, GAGCGACGAGUACAAGAUGTT, CACCCUGCGGAACUUGUUCAATT and CGCCUUUAGACCCAUGGAAGUTT; Bbx, UGGGACUUGCAGGCCUGAUAUTT, CCAGUGGGAGCAAGAAGUUUATT and CUCCCUCAAUAUAGUCCUAUUTT; Smad2, GUGAUAGUGCAAUCUUUGUTT, UGGUGUUCAAUCGCAUACUAUTT and CCUUCAGUGCGAUGCUCAATT; Hbp1, CCCUACCCAAUCUGCCAUAUATT, GGCUAACAGAGUUAGCAAATT and CCAGCUAAGUUCAGAUGUATT; Cdx2, GGACAGAAGAUGAGUGGAATT, GAGAAGGAGUUUCACUUUATT and GCUUGCUGCAGACGCUCAATT; Klf6, GCUUGCUGCAGACGCUCAATT, GACCAAUAGCCUGAACUCUTT and GAUGAGUUGACCAGACACUTT; Sox15, CCUGGCAGUUACACCUCUUCTT, GAUGAAGAGAAGCGACCCUUTT and GACUCUUCCACUCCAUAUAAUTT; Med1, UAAGCUUGUGCGUCAAGUAAUTT, GGCUCUCCAAUCCUUAGAACATTand GUGGCCUAUAACACUCUAAUUTT; Elf5, GCCCUGAGAUACUACUAUAAATT, GGACCGAUCUGUUCAGCAATT and GGAGGUUAGUGUACAAAUUTT; and Hif1A, CCAUGUGACCAUGAGGAAATT, GCAGACCCAGUUACAGAAATT and GCAGGAAUUGGAACAUUAUTT. siRNAs were ordered from Hippobio. The injected embryos were transferred to KSOMaa medium (Millipore) and droplets were covered with mineral oil (Sigma) in a Petri dish (Ibidi) and cultured in a tissue incubator (37 °C and 5% CO2) (Thermo Fisher Scientific). Embryos were collected at the 8cell or blastocyst stage, and single-embryo RNA-seq or immunofluorescence staining was performed to confirm KD or marker gene expression.
scRNA-seq or single-embryo RNA-seq library generation and sequencing
scRNA-seq and single-embryo RNA-seq library preparation were performed using a modified Smart-seq3 protocol51,52. The zona pellucida was gently removed by treating with Tyrode’s solution (Sigma). Isolated single cells or single embryos at the 8cell or blastocyst stage after siRNA microinjection were mouth-pipetted into lysis buffer. Lysis buffer consisted of 0.15% Triton X-100 (VWR Life Science), 5% PEG8000 (Thermo Fisher Scientific), 0,5 μM oligo-dT (Supplementary Table 1), 0.5 mM dNTPs and 0.5 U RNase inhibitor (Takara). After dispensing, lysis tubes were briefly centrifuged to ensure that lysis buffer was located under the overlay. Tubes of sorted cells were denatured at 72 °C for 10 min, followed by the addition of the reverse transcription mix. The reagent concentrations were as follows: 25 mM Tris-HCl pH 8.3 (Sigma), 30 mM NaCl (Sigma), 0.5 mM GTP (Thermo Fisher Scientific), 2.5 mM MgCl2 (Sigma), 8 mM DTT ((Thermo Fisher Scientific), 0.25 U μl–1 RNase inhibitor, 2 μM TSO (5′-AAGCAGTGGTATCAACGCAGAGTACATG(r)G(r)G(+)-3′) and 2 U μl–1 Maxima H Minus reverse transcriptase (Thermo Fisher Scientific). Reverse transcription and template switching were carried out at 42 °C for 90 min followed by 10 cycles of 50 °C for 2 min and 42 °C for 2 min. The reaction was terminated by incubating at 85 °C for 5 min. Indicated volumes of PCR master mix were dispensed, which contained 1× KAPA HiFi PCR buffer (Roche), 0.3 mM dNTPs each (Roche), 0.5 mM MgCl2 (Roche), 0.6 mM P2 primer (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-3′) and 0.2 μM IS primer (5′-AAGCAGTGGTATCAACGCAGAGT-3′) and 0.02 U μl−1 KAPA HiFi DNA polymerase (Roche). Pre-amplification was performed as follows: 3 min at 98 °C for initial denaturation, 16 cycles of 20 s at 98 °C, 30 s at 65°C, and 5 min at 72 °C. Final elongation was performed for 5 min at 72 °C. After PCR, samples were pooled and purified using a TIANquick Mini Purification kit (Tiangen) and 0.8× AMPure XP beads (Beckmann).
After purification, 1 μl cDNA was used for measuring the concentration. About 10 ng cDNA was subjected to tagmentation with 1 μM PAT–MEA in the reaction buffer (10 mM TAPS-NaOH pH 8.3 (Sigma), 5 mM MgCl2 (Sigma), 10% N,N-dimethylformamide (DMF) (Sigma)) at 55 °C for 10 min. Samples were then treated with 0.025% SDS at 55 °C for 10 min and 0.15% Triton X-100 at 37 °C for 10 min. Enrichment PCR was performed as follows: 3 min at 95 °C for initial denaturation, 16 cycles of 20 s at 98 °C, 15 s at 67 °C, and 1 min at 72 °C, and 5 min at 72 °C. The library was purified with 1× AMPure XP beads. Size selection was carried out first with 0.5× AMPure XP beads and second with 0.5× XP AMPure beads in the supernatant to obtain 200–1,000 bp fragments for sequencing. The libraries were sequenced with paired-end 150-bp reads on NovaSeq 6000 platform (Illumina).
Immunofluorescence staining
Injected embryos were fixed with 4% paraformaldehyde (Sigma) for 10–15 min. PBST (PBS + 0.5% Triton-X) was added for 20 min at room temperature to permeabilize the embryos and the samples were subsequently incubated with blocking buffer (PBS + 0.1% Tween-20 + 5% NDS) for 4 h at 4 °C. After blocking, embryos were incubated with SOX2 (Active motif) and CDX2 (BioGenex), diluted in blocking buffer, at 4 °C overnight. Samples were then washed with PBS 3 times and incubated with secondary antibodies (Invitrogen), diluted in blocking buffer, for 2–4 h at room temperature. Finally, blastocysts were incubated with 600 nM DAPI solution (Thermo Fisher) for 5 min at room temperature and were rinsed with PBS before visualization. Images were acquired using a confocal microscope (Zeiss LSM 710).
CRISPRa-mediated TF activation in mouse ES cells
The sgRNAs targeting the promoter of each of the candidate totipotency-related TFs were synthesized and inserted into a CROP-opti vector separately (Addgene, 106280) (Supplementary Table 9). Three libraries of sgRNAs included candidate TFs (CEBPG, LBX1, ETS2, MEF2D, ESR2, ESR1 and ALX1), positive control TFs (ZSCAN4 and DUX) and a non-targeting control as previously described53 at equal molar ratios. The supernatant with lentivirus was collected 18 h after transfection and filtered to remove cell debris. The mouse ES cells were infected (8 μg ml–1 polybrene) with various titres of lentivirus to achieve different multiplicity of infection values. At 24 h after transduction, new culture medium with 2 μg ml–1 puromycin was added for 48 h for selection. Cells after transduction and selection were collected for scRNA-seq. Cell pellets were fixed with 1% formaldehyde at 4 °C for 10 min and were preserved at −80 °C or used immediately. Single-cell RNA-seq for mouse ES cells for capturing both mRNA and sgRNAs was conducted as per the SPLiT-seq pipeline as previously described54.
Data processing
TACIT data were processed as previously described19, but with a few modifications for single cells. Raw TACIT sequencing data were evaluated using FastQC (v.0.11.5), followed by mapping to the mouse reference genome mm10 by Bowtie2 (v.2.2.9)55. Mapped reads with MAPQ vales less than 30 were considered as multi-mapped reads and filtered out using Samtools (v.1.9). PCR duplicates were also removed using Picard (v.2.2.4). For aggregated analysis, single-cell .bam files were merged with Samtools. For peak calling, MACS2 (v.2.1.1)56 with the ‘–broad’ parameter was used to call peaks for aggregated profiles of TACIT data. Raw CoTACIT sequencing data were de-multiplexed and paired using an in-house code as previously described19. Sequencing data for each histone modification was performed according to the analysis pipeline as described for TACIT data.
Correlation analysis for TACIT data
For correlation analysis between different experiments, we calculated the normalized mean scores in 5-kb bins of the genome by using the multiBigwigSummary function in deepTools (v.3.5.1)57. The Spearman correlation or Pearson correlation was calculated between replicates and plotted using the plotCorrelation function.
Genome-coverage analysis
To calculate the genome coverage at each developmental stage, we first called peaks for aggregated .bam files of each histone modification. We used MACS2 to call peaks with parameters of ‘–nolambda–nomodel -q 0.05–broad’. Next, we binned the mm10 genome into 200-bp genomic intervals, and for each histone modification, genome coverage at a specific stage was calculated as the percentage of genome intervals that overlapped with peaks at that stage. To evaluate genome coverage for single cells, the genome was first binned into 200 bp and bins with histone modification signals ≥ 1 were defined as covered bins. The percentage of covered bins was defined as genome coverage for each single cell.
Clustering of TACIT and CoTACIT data
TACIT alignment files were converted to a matrix with genomic intervals (instead of peaks) as rows and cells as columns using cisTopic (v.0.3.0)58. For different histone modifications, different sizes of genomic intervals were used as follows: 5 kb bins for H3K27ac; 10 kb bins for H3K4me3, H3K36me3 and H3K27me3; and 15 kb bins for H3K4me1 and H3K9me3. Clustering of embryonic cells on the basis of histone modifications was performed using Seurat (v.4.3.0). In brief, the cell–bins matrix was first normalized with the term frequency–inverse document frequency (TF–IDF), followed by dimensionality reduction with singular value decomposition (SVD). Next, 2:20 or 1:20 (only for H3K4me3) dimensions were used for identifying clusters and for UMAP visualization. For clustering of CoTACIT data from the early 2cell stage, fragment counts in 5 kb genome windows were used for all three histone modifications. The Seurat (v.4) WNN59 framework was used to generate a multimodal representation using dimensions 1:20 (H3K4me3), 2:20 (H3K27ac) and 2:20 (H3K27me3).
Normalization of Euclidean distance
To evaluate cell heterogeneity among stages for each histone modification, we first calculated the Euclidean distance between each pair of cells in the same stage as shown in the UMAP embeddings. The median Euclidean distance of zygotes was set as the baseline for normalization of other cells across all stages.
Generation of synthetic cells
To investigate the dynamics of chromatin states during mouse pre-implantation development, we generated synthetic cells as follows:
-
1)
We ordered scRNA-seq cells along the developmental trajectory using Monocle3 (ref. 60) and merged five adjacent single cells along pseudotime into one RNA synthetic cell.
-
2)
We integrated H3K4me1, H3K4me3, H3K36me3 and H3K27ac TACIT profiles with gene expression. In brief, the cell–peak or cell–bin matrix for each histone modification was first generated using cisTopic58. The GeneActivity function of Seurat (v.4) was used to create a gene-activity score matrix based on the cell–peak or cell–bin matrix. Next, anchors between the two modalities were identified with the FindTransferAnchors function. In particular, many titrations were performed to obtain the highest prediction score, including using the cell–peak or cell–bin matrix, or the bin size of the cell–bin matrix. TACIT cells with a prediction score lower than 0.2 were filtered out. Notably, for integrating cells in the 2cell stage, histone-modification signals in non-canonical broad binding regions were excluded before Seurat integration (5 kb for H3K4me3 and H3K27ac, 20 kb for H3K36me3 and H3K4me1).
-
3)
We integrated H3K27ac CoTACIT profiles with gene expression in the same way as described in step (2). As H3K27ac, H3K27me3 and H3K9me3 profiles were experimentally linked, we directly transferred corresponding H3K27me3 and H3K9me3 profiles to the linked RNA synthetic cells.
-
4)
Having obtained 155 RNA synthetic cells interpolated with six histone-modification profiles, we performed hierarchical clustering with RNA synthetic cells on the basis of multimodal histone modifications. The number of clusters closely corresponded to the exact cell number of each developmental stage, such as two clusters for the 2cell stage, four clusters for the 4cell stage, and so on. Next, we aggregated histone-modification profiles of cells in the same cluster, which led to 90 synthetic single cells with joint profiles of 6 histone modifications. To reduce effects from sequencing depth, we normalized cell numbers and non-duplicated reads before aggregating data.
WNN analysis for interpolated single cells
After integrating the TACIT and CoTACIT data with the RNA data, we obtained interpolated single cells simultaneously with six histone-modification measurements. We used the Seurat (v.4) WNN framework to generate a multimodal representation of interpolated single cells. We used the FindMultiModalNeighbors function to generate a WNN graph using the following dimensions: H3K4me1, 2:15; H3K4me3, 2:15; H3K27ac, 2:15; H3K36me3, 2:15; H3K27me3, 2:20; and H3K9me3, 2:15.
ChromHMM for synthetic cells
To integrate the six histone modification profiles, we used the multivariate HMM introduced in ChromHMM61. We binarized all .bam files for each synthetic cell using the binarizeBam function of ChromHMM with default parameters. We used the LearnModel function with default parameters to learn 12 states separately on each synthetic cell. To reduce noise and mitochondrial interference, all reads from mitochondrial DNA are filtered out. Next, we annotated each state in three steps: (1) filtering out chromatin states with extremely low genome coverage (<0.001%), because these were probably from technical noise; (2) defining hidden chromatin states based on the combination of histone modifications; and (3) correcting the annotation on the basis of the overlap in the main genome categories. Finally, we labelled the 12 states as multivalent (all histone modifications), weak promoters (H3K4me3), strong promoters (H3K4me3 and H3K27ac), weak enhancers (H3K4me1), strong enhancers (H3K4me1 and H3K27ac), poised gene bodies (H3K36me3 and repressive histone modifications), active gene bodies (H3K36me3 and active histone modifications), polycomb-protein-associated heterochromatin (only H3K27me3), H3K9me3-associated heterochromatin (only H3K9me3), heterochromatin (H3K27me3 and H3K9me3), and quiescent/low.
scChromHMM for interpolated cells
To annotate chromatin states at single-cell resolution for blastocyst cells, we first generated single-cell profiles with simultaneous measurements of six histone modifications. As described above, we integrated H3K4me1, H3K4me3, H3K27ac and H3K36me3 TACIT profiles as well as H3K27ac CoTACIT profiles with gene expression. In addition, we annotated ICM and TE cells on the basis of the expression of ICM or TE marker genes.
We used the LearnModel command of ChromHMM46 to train a 12-state model with aggregate ICM and TE profiles. Next, we ran the forward–backward algorithm to learn the posterior probability distribution for interpolated single cells. We set up the bin size of 2,000 bp and grouped the states into 6 categories (enhancers, promoters, gene bodies, polycomb-protein-associated heterochromatin, H3K9me3-associated heterochromatin and quiescent/low). We also merged five adjacent single cells along pseudotime52.
Clustering by scChromHMM-defined chromatin-state annotations of synthetic single cells
For clustering cells on the basis of scChromHMM-defined chromatin-state annotations of all genomic intervals (Fig. 4a), we used the posterior probability matrix for each state as input for TF–IDF normalization, SVD dimensionality reduction, cluster finding and UMAP visualization with Seurat (v.4). We used 1:5 dimensions for clustering and visualization. For clustering cells on the basis of chromatin-states annotations in all TSSs, we selected genomic intervals that were ±2 kb flanking TSS regions and averaged the posterior probability for a specific chromatin state in each TSS as the average probability of this chromatin state. Next, we used the mean probability matrix for each chromatin state for TF–IDF normalization, SVD dimensionality reduction, cluster finding and UMAP visualization. Dimensions 1:5 were used for clustering and visualization.
Identification of differential bins between totipotent and pluripotent cells
In Extended Data Fig. 10b, for each chromatin-state labelling, after TF–IDF normalization, SVD dimensionality reduction and cluster finding (as described above), we used the FindMakers function in Seurat (v.4) to find differential bins between cells at the 2cell and 8cell stage. Next, to quantify the establishment of totipotency-related chromatin regions during development, we calculated the percentage of differential bins that were already annotated as corresponding chromatin states for each synthetic cell.
Identification of feature classifier bins for defining totipotency
We adopted two strategies for identifying putative totipotency-related classifier bins between 2cell 1 versus 2cell 2 (strategy 1) and 2cell versus 8cell (strategy 2), in which the former aimed to rule out differences in stage. For each strategy, a cell–bin probability matrix was first generated for each chromatin state, and genomic regions for which posterior probability exhibited a correlation of more than 80% or less than −50% with the expression of totipotency marker genes was selected. Next, all highly correlated candidate bins were aggregated to generate the state matrix with state annotation information for each genomic interval. The state matrix of synthetic cells at the 2cell and 8cell stages was used as input for constructing the random forest training model62, with labels as ‘toti-high’ and ‘toti-no’ groups, respectively.
Identification of feature classifier bins for ICM and TE specification
With the scChromHMM annotation for blastocyst synthetic cells, we used 29 synthetic cells as training cells to build a random forest machine-learning model and 10 synthetic cells as testing cells for cross-validation. This trained random forest model prioritized 780 genomic intervals for which chromatin states are essential for the first lineage specification and predicted the ICM or TE tendency of 4cell, 8cell and morula cells.
TF motif analysis
In Extended Data Fig. 7g, we intersected our ChromHMM-annotated regions with ATAC-seq peaks and called motifs on these ChromHMM–ATAC regions using Homer. By default, Homer uses random genomic regions as the background. For motif analysis in Figs. 4e and 5e, we directly used classifier bins for calling motifs because the bin size was 200 bp and 2,000 bp, respectively. We considered that such bins are sufficiently narrow for calling TF motifs. To disentangle the influence of open chromatin during TF motif enrichment assessment in classifier bins, previously published ATAC-seq data (GSE66390)18 were used. All open-chromatin regions from the 2cell and 8cell stages were combined and used as the background for Fig. 4e. ATAC-seq peaks were segmented into 200-bp bins, matching the classifier bin size. First, 765 TF motifs were identified on classifier bins that were annotated as active states (promoter, enhancer and gene bodies) for all synthetic cells. Next, we selected 327 TF motifs that were highly enriched in 2cell 1 or 2cell 2 (–log10(P) > 8) and depleted in 8cell (–log10(P) < 2). Finally, only TF motifs with detected expression in 2cell 1 or 2cell 2 (TPM > 2) were chosen as putative totipotency-related TFs, as listed in Supplementary Table 8. In Fig. 5e, TF motif enrichment was calculated using Homer for classifier bins that were annotated as enhancers for each synthetic cell. Open-chromatin regions from the 4cell to blastocyst stages were used as the background, with ATAC-seq peaks binned to 2,000 bp, matching the classifier bin size. Only synthetic cells defined as ICM-potential or TE-potential were used for TF motif enrichment analysis during ICM or TE specification, respectively. We enriched 59 TF motifs on classifier bins that were annotated as active states for ICM-potential cells and 42 for TE-potential cells (–log10(P) > 5). Next, we selected eight putative ICM-related TFs and five putative TE-related TFs on the basis of expression levels (RPKM > 1.5 in Ribo-lite data63).
Distribution of classifier bins
The enrichment of the classifier bins in Extended Data Fig. 10c was calculated using observed versus expected probability as previously described12. The observed probability was calculated using the length of classifier bins covering the related genomic regions versus the length of the total classifier bins, and the expected probability was calculated using the length of the total related genomic regions versus the length of the mouse genome. Promoter was defined as ±1 kb genomic region around all TSSs. The locations of annotated repeats (RepeatMasker) were downloaded from the UCSC Genome browser18,64.
Gene expression with chromatin states
For each synthetic cell, the median gene expression was presented from cells belonging to the same synthetic cell. Each chromatin region was linked to the nearest genes using Homer, and expression for all genes and all samples were then combined and split by categories of chromatin states. A boxplot was plotted for each chromatin state. To eliminate the effects of non-canonical chromatin-binding features, only synthetic cells at the 2cell and 4cell stages were included in this analysis.
Identifying promoter–enhancer pairs
To identify promoter–enhancer pairs, we used TSS-proximal signals (± 5 kb flanking TSSs) to build a peak–cell matrix for H3K4me3 and used TSS-distal signals to build a peak–cell matrix for H3K27ac. Next, we integrated these two matrices and evaluated co-occurrence of pairs with Cicero (v.1.14.0)65. We also established a criterion in which only pairs that link a H3K4me3 peak and a H3K27ac peak were defined as promoter–enhancer pairs. Overall, we identified 43,983 promoter–enhancer pairs (Cicero score, S > 0.1). To find putative functional promoter–enhancer pairs implicated in ZGA, we chose pairs for which H3K4me3 peaks fell within ±2 kb flanking the TSS of genes that are activated after ZGA66. We calculated the fold change (FC) of the Cicero score between 2cell 1 and 2cell 2 clusters (FC = S2cell 2/S2cell 1). We defined 2cell 1-specific (FC < 0.25, S2cell 1 > 0.3, and S2cell 2 < 0.05), 2cell 2-specific (FC > 2, S2cell 1 < 0.05, and S2cell 2 > 0.3) and shared (S2cell 1 > 0.3, and S2cell 2 > 0.3) promoter–enhancer pairs. Finally, we identified 515 2cell 1-specific, 1,138 2cell 2-specific and 159 shared promoter–enhancer pairs.
Similarly, we chose promoter–enhancer pairs for which H3K4me3 peaks or H3K27ac peaks fell into copies of MERVL elements as MERVL-associated pairs, whereby MERVLs function as promoters or enhancers, respectively. We identified 332 enhancer–promoter pairs in which MERVL elements functioned as promoters, and 866 enhancer–promoter pairs in which MERVL elements functioned as enhancers.
Re-analysis of Hi-C data
The allvalidPairs matrix for the late 2cell stage was downloaded from the GEO database (accession number GSE82185)67. To identify interactions, we used the analyzeHiC function of Homer (v.4.11)68 at 50 kb resolution and plotted interactions with Python (v.3.9.7).
Chromatin states and expression of transposable elements
For the enrichment analysis in Fig. 4f, the observed probability was calculated using the length of classifier bins covering the related transposable elements versus the length of the total classifier bins, and the expected probability was calculated using the length of the total related transposable elements versus the length of the mouse genome. Overall, we identified 75 transposable elements that were highly enriched (log2(overexpression) > 1) in 2,583 totipotency-related classifier bins. For the annotation of chromatin states of the enriched 75 transposable elements in Fig. 4g, we calculated the percentage of transposable element copies that were defined as promoters, enhancers, gene bodies, heterochromatin or quiescent/low regions for each synthetic cell. To quantify expression levels of these transposable elements, we mapped raw scRNA-seq reads to the mm10 genome using Hisat2 (v.2.2.1)69 and filtered out mapped reads with MAPQ less than 10 with Samtools (v.1.9). We calculated the numbers of TPM based on the locations of annotated repeats (RepeatMasker) downloaded from the UCSC Genome browser.
Analyses for multiplexability of CRISPRa in mouse ES cells
To evaluate the efficacy of totipotency activation of our CRISPRa experiments in mouse ES cells, we first quantified the abundance of designed sgRNAs targeting candidate TFs (CEBPG, LBX1, ETS2, MEF2D, ESR2, ESR1 and ALX1), positive control TFs (ZSCAN4 and DUX), and non-targeting control based on detected sgRNA unique molecular identifiers (UMIs). sgRNAs with fewer than 16 UMIs were filtered out for further analyses. Perturbed cells were clustered and projected in UMAP together with totipotent blastomere-like cells and pluripotent stem cells in public scRNA-seq datasets42 using Seurat (v.4). To analyse the perturbation effects of candidate TF genes, we ranked the genes and calculated perturbation correlation among of them based on cells receiving only one sgRNA using MUSIC70 with default parameters. Furthermore, the totipotency score for cells with each combination of gene perturbation was calculated on the basis of the totipotent gene signature.
RNA-seq analysis for single embryos with siRNA knockdown
Sequenced reads with adaptor and low-quality bases were removed. The clean reads were aligned to the mm10 reference genome (RNA library) and in-house siRNA database (siRNA library) with Hisat2. For each single embryo, the expression level of a gene was normalized by the TPM. The KD information for each embryo at the 8cell and blastocyst stage as metadata was incorporated together with the gene-expression matrix in Seurat analysis. The individual embryos were visualized by UMAP using Seurat (v.4) with default parameters. The TF activity for cells with TE or ICM candidate gene KD was evaluated using SCENIC71.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequencing data have been deposited into the GEO database with accession numbers GSE235109 (TACIT data) and GSE259393 (CoTACIT, RNA-seq, embryo-barcoded TACIT and itChIP data). Other public datasets used in this study were downloaded from the GEO database with the following accession numbers: scRNA-seq (GSE45719), sc-itChIP-seq (GSE109757), CoBATCH (GSE129335), scCUT&Tag (GSE124557), two droplet-based scCUT&Tag datasets (GSM5034344 and GSE157637), bulk H3K4me3 ChIP–seq in mouse ES cells (GSM1000124), bulk H3K27ac ChIP–seq in mouse ES cells (GSM1000126), bulk H3K36me3 ChIP-seq in mouse ES cells (GSM1000109), bulk H3K27me3 ChIP-seq in mouse ES cells (GSM1000089), bulk H3K4me3 ChIP–seq in embryos (GSE71434), bulk H3K27ac ChIP–seq in embryos (GSE72784), bulk H3K36me3 ChIP–seq in embryos (GSE112835), bulk H3K27me3 ChIP–seq in embryos (GSE76687), bulk H3K9me3 ChIP–seq in embryos (GSE97778), bulk H2A.Z ChIP–seq in mouse embryonic fibroblasts (GSE51579), bulk ATAC-seq in embryos (GSE66390), bulk Hi-C in embryos (GSE82185) and mouse reference genome mm10 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001635.20/). Source data are provided with this paper.
Code availability
Custom scripts used in this study are available from GitHub (https://github.com/Helab-bioinformatics/TACIT).
References
Burton, A. & Torres-Padilla, M. E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–734 (2014).
Zhang, Y. & Xie, W. Building the genome architecture during the maternal to zygotic transition. Curr. Opin. Genet. Dev. 72, 91–100 (2022).
Hemberger, M., Dean, W. & Reik, W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 10, 526–537 (2009).
Zernicka-Goetz, M., Morris, S. A. & Bruce, A. W. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467–477 (2009).
Posfai, E. et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 23, 49–60 (2021).
Krawczyk, K., Kosyl, E., Czescik-Lysyszyn, K., Wyszomirski, T. & Maleszewski, M. Developmental capacity is unevenly distributed among single blastomeres of 2-cell and 4-cell stage mouse embryos. Sci. Rep. 11, 21422 (2021).
Junyent, S. et al. The first two blastomeres contribute unequally to the human embryo. Cell 187, 2838–2854 (2024).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016).
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Zheng, H. et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016).
Wang, C. et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018).
Yu, H. et al. Dynamic reprogramming of H3K9me3 at hominoid-specific retrotransposons during human preimplantation development. Cell Stem Cell 29, 1031–1050 (2022).
Xu, R. et al. Stage-specific H3K9me3 occupancy ensures retrotransposon silencing in human pre-implantation embryos. Cell Stem Cell 29, 1051–1066 (2022).
Burton, A. et al. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol. 22, 767–778 (2020).
Guo, F. et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 (2017).
Wang, Y. et al. Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat. Commun. 12, 1247 (2021).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216 (2019).
Ai, S. et al. Profiling chromatin states using single-cell itChIP–seq. Nat. Cell Biol. 21, 1164–1172 (2019).
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 39, 825–835 (2021).
Wu, S. J. et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 39, 819–824 (2021).
Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10, 3747 (2019).
Martire, S. & Banaszynski, L. A. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 21, 522–541 (2020).
Boix, C. A., James, B. T., Park, Y. P., Meuleman, W. & Kellis, M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 590, 300–307 (2021).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Creyghton, M. P. et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 (2008).
Long, H. et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576–581 (2020).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Park, S. J. et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 27, 2736–2748 (2013).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Cree, L. M. et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 40, 249–254 (2008).
Deng, Q., Ramskold, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Macfarlan, T. S. et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012).
Hendrickson, P. G. et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934 (2017).
Zhang, W. et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 47, 8485–8501 (2019).
Shen, B. et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182, 59–72 (2020).
Timmons, J. A., Szkop, K. J. & Gallagher, I. J. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol. 16, 186 (2015).
Shen, H. et al. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 184, 2843–2859 (2021).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017).
Goolam, M. et al. Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165, 61–74 (2016).
Fasching, L. et al. Early developmental asymmetries in cell lineage trees in living individuals. Science 371, 1245–1248 (2021).
Zhang, B. et al. Characterizing cellular heterogeneity in chromatin state with scCUT&Tag-pro. Nat. Biotechnol. 40, 1220–1230 (2022).
Baran, Y. et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 20, 206 (2019).
Chan, C. J. et al. Hydraulic control of mammalian embryo size and cell fate. Nature 571, 112–116 (2019).
Zhang, Y. et al. Genetic reporter for live tracing fluid flow forces during cell fate segregation in mouse blastocyst development. Cell Stem Cell 30, 1110–1123 (2023).
Strnad, P. et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 (2016).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Xiong, H., Luo, Y., Wang, Q., Yu, X. & He, A. Single-cell joint detection of chromatin occupancy and transcriptome enables higher-dimensional epigenomic reconstructions. Nat. Methods 18, 652–660 (2021).
Horlbeck, M. A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Langdon, W. B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8, 1 (2015).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Bravo Gonzalez-Blas, C. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Ernst, J. & Kellis, M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017).
O’Huallachain, M. et al. Ultra-high throughput single-cell analysis of proteins and RNAs by split-pool synthesis. Commun Biol. 3, 213 (2020).
Xiong, Z. et al. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat. Cell Biol. 24, 968–980 (2022).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics https://doi.org/10.1002/0471250953.bi0410s25 (2009).
Pliner, H. A. et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871 (2018).
Adam, R. C. et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015).
Du, Z. et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Duan, B. et al. Model-based understanding of single-cell CRISPR screening. Nat. Commun. 10, 2233 (2019).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Acknowledgements
We thank all members of the He Laboratory for comments on this article; members of the F. Tang and W. Xie Laboratories and Y. Wei for sharing protocols and providing technical support; staff at the Laboratory Animal Research Center at Peking University for assistance with animal experiments; staff at the Confocal LSM 710 Core at the National Center for Protein Sciences at Peking University for technical help; and staff at the National Center for Protein Sciences at Peking University for help with the MGI2000 NGS platform. Part of the analyses was performed using the High Performance Computing Platform of the Center for Life Sciences, Peking University. A.H. was supported by grants from the National Basic Research Program of China (2021YFA1100100), the National Natural Science Foundation of China (32025015 and 32192401) and the Peking–Tsinghua Center for Life Sciences. Y.Y. was supported by grants from the National Key R&D Program of China (2022YFA1106200) and the National Natural Science Foundation of China (32200683 and 32370873).
Author information
Authors and Affiliations
Contributions
A.H. conceived and designed the study. M.L. designed and performed TACIT, CoTACIT and scRNA-seq experiments supervised by A.H. M.L. and Y.Y. conducted embryo-barcoded TACIT experiments. Y.Y. and M.S. performed siRNA KD experiments. C.D. and K.X. contributed to embryo collection experiments and reagents. M.L. and X.C. performed computational analyses supervised by A.H. H.X. contributed to CRISPRa data analyses supervised by A.H. M.L., X.C., K.T. and Y.L. performed machine-learning analyses supervised by Q.C.Z. and A.H. M.L. and A.H. wrote the paper. All authors participated in data discussion and interpretation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Optimization of key steps in TACIT and data quality in mouse embryonic stem cells.
(a-b) Optimization of critical conditions for reverse crosslinking. Based on yielding DNA fragments of <1 kb (a) and non-duplicated reads per cell (b), the condition of incubation at 55 °C for 15 min with 0.1 mg/ml Proteinase K is chosen in TACIT experiments. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). For gel source data, see Supplementary Fig. 1. The cell number for each group in (b) is 96. (c) Left, bioanalyzer DNA analysis of one example of TACIT libraries. Right, fragment length distribution of one example of TACIT libraries. (d) Track views showing H3K4me3, H3K27ac, H3K36me3, and H3K27me3 TACIT signals on representative locus in mouse embryonic stem cells (mESCs). ENCODE bulk data for H3K4me3, H3K27ac, H3K36me3, H3K27me3 ChIP-seq were downloaded from GSM1000124, GSM1000126, GSM1000109, and GSM1000089, respectively. (e) Hierarchical clustering of the aggregate TACIT single-cell profiling of different histone modifications using signals in peak regions. (f) Violin plot showing the fraction of reads in peak (FRiP) for single cell TACIT data, with simulated random profiling as control. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). The cell number for each group is below: H3K4me3 (94), H3K27ac (94), H3K36me3 (93), H3K27me3 (92), and random (96). (g) Distribution of non-duplicated reads for single cells in TACIT, sc-itChIP-seq, CoBATCH, scCUT&Tag, and droplet-based scCUT&Tag datasets. TACIT, sc-itChIP-seq, and CoBATCH datasets are derived from mouse embryonic stem cells, scCUT&Tag dataset is from K562 cells, and the two droplet-based scCUT&Tag datasets are from human peripheral blood mononuclear cells and mouse brain cells, respectively. sc-itChIP-seq (n = 1,903), CoBATCH (n = 2,161), scCUT&Tag (n = 217), and two droplet-based scCUT&Tag datasets (n = 133,696 and 2,028) were downloaded from GSE124557, GSE129335, GSE124557, GSM5034344, and GSE157637 respectively. Cells with fewer than 100 non-duplicated reads in droplet-based scCUT&Tag datasets were filtered out. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles).
Extended Data Fig. 2 TACIT experiments with mouse embryos.
(a) Representative images of embryos used for TACIT library preparation. The two pronucleus of the zygote are indicated with the white arrows. Shown are representative images from three biological replicates. Scale bars, 20 μm. (b) Representative images of tagemented single cells before reverse crosslinking and lysis. Shown are representative images from three biological replicates. Scale bars, 50 μm. (c) Exemplification of scatter plots showing Pearson correlation coefficient for both technical replicates (H3K27me3) and biological replicates (H3K27ac) in mouse embryos. Technical replicates represent two TACIT libraries generated from the same batch of embryos, while biological replicates represent two independent TACIT experiments at the matching stages. The correlation is calculated based on aggregate TACIT signals in 5-kb bins of the genome. (d) Pearson correlation between aggregated H3K4me3, H3K27ac, and H3K9me3 TACIT profiles and corresponding low-input ChIP-seq data. The correlation is calculated based on signals in genome-wide 10-kb or 20-kb bins. (e) Track views displaying aggregate histone modification signals on representative loci. Active histone modifications, such as H3K4me1, H3K4me3, H3K27ac, and H3K36me3, exhibit non-canonical, broad binding patterns before ZGA. (f) Scatter plots showing Pearson correlation coefficient between aggregated H3K36me3 signals of TACIT and itChIP for zygote and 2-cell stage. The correlation is calculated based on signals in genome-wide 20-kb or 50-kb bins. (g) Receiver operating characteristics curves for H3K36me3 TACIT data. Peaks of itChIP-seq data were used as the gold standard (15,069 for zygote, 12,035 for 2-cell). (h) Genome coverage for aggregate profiles. For each histone modification, genome coverage is defined as the proportion of the genome covered by peaks for aggregate cells each stage. (i) Median genome coverage per cell in TACIT experiments. To evaluate H3K4me1 genome coverage for single cells, the genome was firstly binned into 200 bp and bins with H3K4me1 signals≥1 were defined as H3K4me1 covered bins. Similar analyses were applied for H3K4me3, H3K27ac, H3K36me3, H3K27me3, H3K9me3, and H2A.Z.
Extended Data Fig. 3 UMAP embedding of TACIT data.
(a-b) UMAP visualization of all TACIT profiles for each developmental stage based on histone modification signals either genome-wide (a) or within peaks (b). Cell-to-cell variability within a given stage increases gradually as embryos develop. Heterogeneities within zygotic cells may arise likely because these cells come from different pronuclear stages (PN0/1 to PN5). For blastocysts, ICM and TE cells are annotated manually based on histone modification signals around ICM and TE feature genes.
Extended Data Fig. 4 Data quality of CoTACIT with mouse embryos.
(a) Top, bioanalyser and tapestation plots of one example of CoTACIT libraries. Bottom, fragment lengths distribution of one example. (b) Track view displaying aggregate CoTACIT profiles of different histone modifications on representative loci. (c) Hierarchical clustering of the aggregate TACIT, CoTACIT, and corresponding low-input ChIP-seq data using signals in aggregate peaks or genome-wide 20-kb bins.
Extended Data Fig. 5 Quality controls in generation of synthetic cells.
(a) Combined UMAP visualization of the scRNA-seq dataset from this study (n = 1012) and data from by Deng et al.35. Each dot represents an individual cell and is colored by technologies (left) or stages (right). (b) Heatmap showing the Spearman correlation coefficients between H3K4me3 TACIT cells and RNA synthetic cells at each developmental stage. (c) Violin plots showing Spearman correlation coefficients between H3K27ac profiles from TACIT and CoTACIT that are linked to the same scRNA-seq synthetic cells after integration analysis. Box plots centre lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× IQR.The TACIT cell number for each stage is below: 2-cell (66), 4-cell (72), 8-cell (89), morula (121), and blastocyst (166). The CoTACIT cell number for each stage is below: 2-cell (52), 4-cell (70), 8-cell (118), morula (190), and blastocyst (197). (d) UMAP visualization of interpolated single cells on the basis of histone modifications. (e) Hierarchical clustering of synthetic cells from 2-cell, 4-cell, 8-cell, morula and blastocyst embryos. (f) Heatmap showing Pearson correlation of epigenetic profiles for synthetic cells at zygote, 2-cell, 4-cell and 8-cell stages. Correlation coefficients are labeled on the plots. (g) Track view showing aggregated TACIT data of all six histone modifications in 2cell_2 and 4cell_4. (h) Scatter plots showing Spearman correlation between H3K4me3 and H3K27ac signals in synthetic cells at 2-cell and 4-cell stages. Histone modification signals were calculated in ±50 kb regions flanking the TSS of genes exclusively expressed in early-stage embryos. (i) Comparison between H3K27ac and H3K27me3 profiles based on genome-wide distribution for synthetic cells across different developmental stages. Each line represents one synthetic cell. (j) Comparison between H3K27ac and H3K27me3 profiles based on signals in peak regions for synthetic cells across different developmental stages. Each line represents one synthetic cell.
Extended Data Fig. 6 Characterization of feature chromatin states during mouse preimplantation development.
(a) Annotation of genic and non-genic elements for each ChromHMM-trainned chromatin state. Chromatin state definitions are labeled on the left for each synthetic cell. (b) Emission probabilities for random control synthetic cells by single-cell ChromHMM. Random control synthetic cells are generated via randomly merging interpolated cells, rather than based on hierarchical clustering. Chromatin state definitions (left) and genome coverage (right) for each state are annotated. (c) Violin plots displaying histone modifications on multivalent regions for all single cells at the zygotic stage. Histone modifications are also calculated in a list of random regions at the same length as multivalent regions, served as a control. (d) Genomic annotation of multivalent regions. (e) Sankey diagram showing chromatin state dynamics of multivalent regions. Each line represents a 200-bp bin categorized by ChromHMM. Multi, multivalent; Pr-W, promoter (weak); Pr-s, promoter (strong); En-W, enhancer (weak); En-S, enhancer (strong); Ge-P, gene body (poised); Ge-A, gene body (active); He-P, heterochromatin (polycomb); He-K9, heterochromatin (H3K9me3); He, heterochromatin (K27 + K9); Quies, quiescent/low. (f) Emission probabilities for GV-oocyte, Zygote, and 2-cell by ChromHMM. Low-input ChIP-seq profiles are either downloaded from GSE76687, GSE71434, GSE207222, GSE112834, GSE98149, or obtained with itChIP-seq in this work. (g) TF motifs identified from chromatin regions annotated as promoters, enhancers or gene bodies for synthetic cells. Representative known regulators for totipotency and pluripotency are shown. Chromatin regions annotated as promoters, enhancers or gene bodies were intersected with ATAC-seq peaks to refine the regions for TF motif enrichment analysis via Homer. P value, one-sided binomial test.
Extended Data Fig. 7 Early 2-cell embryo CoTACIT data facilitates identification of promoter-enhancer pairs.
(a) Boxplots displaying the average expression of ZGA related genes for single cells across developmental stages. Box plots center lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× IQR. The cell number for each stage is below: zygote (95), early 2-cell (59), late 2-cell (77), 4-cell (164), 8-cell (120), morula (251), early blastocyst (167), and late blastocyst (79). (b) Non-duplicated reads per cell in CoTACIT experiments (n = 89). The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). (c) Spearman correlation of aggregated profiles of CoTACIT, TACIT, and published datasets in genome-wide 10-kb bin. (d) Track view displaying aggregate CoTACIT and TACIT signals of different histone modifications on representative loci at the 2-cell stage. (e) Carmer’s V similarity between targeted combinations in promoter, gene body, distal, LINE, SINE, and LTR regions. Promoter regions are defined as the ± 1 kb genomic regions flanking all transcription start sites (TSS) in mm10 genome. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). P value, two-sided Kruskal-Wallis test. All 89 CoTACIT single-cell profiles are used for this analysis. (f) UMAP projection of H3K27ac for cells from zygote to Late-2-cell stages. Cells are colored by developmental stages. TACIT (n = 90) and CoTACIT (n = 89) profiles are both used for this analysis. (g) UMAP projection of RNA for cells from zygote to Late-2-cell stages. Cells are colored by developmental stages. (h) Schematic for breadth score definition. (i) Violin plots showing the distribution of H3K4me3 and H3K27ac breadth scores for single cells. The boxes in violin plots indicate upper and lower quartiles (25th and 75th percentiles). The H3K4me3 TACIT cell number for each stage is below: zygote (20), 2-cell (24), 4-cell (72), 8-cell (88), morula (192), and blastocyst (239). The H3K27ac TACIT cell number for each stage is below: zygote (24), 2-cell (66), 4-cell (72), 8-cell (89), morula (121), and blastocyst (166). (j) Left, heatmap of H3K4me3, H3K27ac, and H3K27me3 signals around ZGA related genes (gene body±10 kb). Rows are genes and columns cells, with cells ordered along the pseudotime. Genes are grouped using k-means clustering based on H3K4me3 and H3K27ac signals. Right, aggregate curves showing dynamics of median H3K4me3, H3K27ac, and H3K27me3 signals along the pseudotime. (k) Schematic of identifying potential promoter-enhancer pairs with Cicero. TSS-proximal H3K4me3 signals and distal-confined H3K27ac signals for each single cell are merged and Cicero is used to identify co-occurrence of H3K4me3 and H3K27ac signals in cells within 2cell_1 or 2cell_2 clusters. Only pairs linking H3K4me3 and H3K27ac peaks are defined as promoter-enhancer pairs and used for following analysis. (l) Distribution of the number of Cicero-linked promoter-enhancer pairs for promoters (left) or enhancers (right). (m) Left, Z-score heatmap showing H3K4me3, H3K27ac, H3K27me3 signals (gene body±10 kb), gene expression and Cicero defined co-binding score of identified ZGA related promoter-enhancer pairs. Rows are promoter-enhancer pairs and columns cells, ordered by pseudotime. Promoter-enhancer pairs are grouped using k-means clustering based on H3K4me3 and H3K27ac signals. Right, aggregate curves showing median RNA, H3K4me3, and H3K27ac signals along the pseudotime. (n) Left, heatmap of H3K4me3, H3K27ac signals, and gene expression for MERVL related promoter-enhancer pairs. Rows are promoter-enhancer pairs and columns cells, with cells ordered along the pseudotime. Promoter-enhancer pairs are grouped using k-means clustering based on H3K4me3 and H3K27ac signals. Right, aggregate curves showing median RNA, H3K4me3, and H3K27ac signals along the pseudotime. (o) Examples of inferred promoter-enhancer pairs, in which MERVL functions as promoters (named as MERVL-promoter, top) or as enhancers (named as MERVL-enhancer, bottom) in 2cell_1 or 2cell_2 clusters. Loops identified in public HiC data are shown underneath. (p) Scatter plots showing the enrichment and expression of transcription factors (TFs) enriched in MERVL-promoter paired enhancers. TFs expressed at the 2-cell stage (FPKM ≥ 2) and with motif enrichment P value < 1 × 10 − 3 are denoted. P value, one-sided binomial test. (q) GO term enrichment (Biological Processes) of MERVL-enhancer linked genes that are highly expressed in 2cell_1 (low ZGA) (left) or highly expressed in 2cell_2 (high ZGA) (right). Top 5 enriched GO terms are shown. P value, one-sided binomial test.
Extended Data Fig. 8 identification and in vitro functional validation of totipotency-related TFs in totipotency induction.
(a) Clustering synthetic cells by posterior probability of different chromatin states in transcription start site (TSS) regions. (b) Progressive changes in expression of totipotency related genes (left) or progressive establishment of totipotency-related chromatin states (right) from zygote to 8-cell developmental stages. The median expression of totipotency related genes is calculated for each synthetic cell (black line, RNA). (c) Bar charts showing enrichment of 2,583 classifier bins in promoter, gene body, distal, and repeat regions. Promoter is defined as the ± 1 kb genomic region around TSS. (d) Overlap of the active cluster of totipotency-related classifier bins (see Fig. 4e) with totipotency feature genes. The totipotency gene list was downloaded from Hui Shen et al. 2021. (e) Bar plots showing GO terms enriched in 120 totipotency-related TFs (listed in Supplementary Table 8). P value, one-sided binomial test. (f) Venn diagram showing overlap of TF motifs enriched on totipotency-related classifier bins generated from the two machine learning models. Both known totipotency-related TF motifs (ZSCAN4 and NR5A2) and newly identified totipotency-potential TF motifs (MEF2D, LBX1, ESR1, ETS1, CEBPG) are shown. P value, one-sided hypergeometric test. (g) Schematic for CRISPRa experiment in mESCs. (h) Scatter plot showing the sgRNA UMI fraction within a given cell (x axis) as a function of the log2 (UMI per sgRNA) received in that cell (y axis). Each dot represents an individual sgRNA species in a specific cell. sgRNAs with less than 16 UMI were not considered for further analyses. (i) Data quality control for detected sgRNA species per cell. (j) UMAP showing the individual cells in this study (n = 10,178) and public scRNA-seq datasets. Public scRNA-seq data were downloaded from GSM5195024.TBLCs, totipotent blastomere-like cells. PSCs, pluripotent stem cells. (k) Pseudotime analysis for individual cells shown in (c). (l) Expression level of totipotency gene signature along the pseudotime of totipotency activation. (m) Cell distribution along the pseudotime of with the increasing of cell totipotency. (n) The proportion of cells receiving different kinds of sgRNAs for pluripotent, intermediate, and TBLCs. NTC, non-targeting control. PC, positive control with DUX and ZSCAN4. (o) The overall perturbation effect ranking lists identified by MUSIC (Duan et al.70, PMID: 31110232) using the cells taking only one sgRNA in the CRISPRa experiment. (p) Heatmap showing the gene–gene perturbation relationships for candidate TFs and positive control TFs (ZSCAN4 and DUX). (q) Violin plot showing the scaled totipotency score for cells with different combination of gene perturbarions. Box plots center lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× IQR. The cell number for each of the eight perturbation combinations is as follows: 1,946, 3,734, 746, 1,068, 651, 1,119, 800, and 1,588. (r) Projection of cells with different combination of gene perturbarions shown in (j) along pseudotime. The cell with median pseudotime distance was labeled by red for each group.
Extended Data Fig. 9 scChromHMM defined feature chromatin states related to ICM and TE specification.
(a) Top, UMAP projection of gene expression of blastocysts (n = 246). Cells are defined as ICM or TE cells based on expression of ICM or TE marker genes. Bottom, expression of representative marker genes of ICM (Klf4, Etv5, Utf1, and Pou5f1) or TE (Cdx2, Id2, Gata3, and Krt18) overlaied on single cell RNA UMAP. (b) Examples of track views on representative gene loci with ChromHMM defined chromatin stages. Top: Aggregate profiles of six histone modifications in ICM and TE cells. Bottom: ChromHMM annotations. (c) Spearman correlation bewteen aggregated ICM and TE H3K4me3, H3K37me3, and H3K9me3 TACIT profiles and corresponding low-input ChIP-seq data. The correlation is calculated based on signals in genome-wide 50-kb bins. (d) Emission probabilities for histone modifications in 12 ChromHMM chromatin states, with labels on the right. (e) Receiver operating characteristic (ROC) of the random forest model. Bam files without one of the six histone modifications are used to construct the random forest model. (f) Genomic distribution of the 780 classifier bins relative to TSS. (g) Enriched GO terms of the Mouse Phenotype Single KO items for the 780 classifier bins. P value, one-sided binomial test. (h) The average Cramer’V similarity between each interpolated cell and ICM or TE cells. Interpolated cells of higher similarity to ICM or TE cells are categorized as ICM-potential or TE-potential cells, respectively. Zygote, 2cell_1, 2cell_2, 4cell_2, 8cell_1, and 8cell_2 exhibit comparable similarity to both cell types, indicating an undefined cellular potential. (i) Bar plots showing GO terms enriched in 59 ICM-related TFs (top) and 42 TE-related TFs (bottom). P value, one-sided binomial test.
Extended Data Fig. 10 Loss-of-function validation of potential TE and ICM regulators.
(a) Bar charts showing the expression of target TFs in both NC and experimental groups of embryos based on single-embryo RNA-seq. P value was determined by two-side t-test. Error bars represent median values ± SEM. The embryo number for each group in 8 C is as below: NANOG KD (20), ZFX KD (15), YY2 KD (26), TCF12 KD (19), CEBPB KD (19), BBX KD (14), SMAD2 KD (15), HBP1 KD (16), CDX KD (12), SOX15 KD (14), MED1 KD (10), NC (19). The embryo number for each group in blastocyst is as below: HNF4A (19), KLF6 KD (10), ELF5 KD (20), HIF1A KD (13), and NC (15). (b) Stereomicroscopic representative images of embryos at the indicated time points with different siRNAs. Representative images out of three independent experiments are shown. Scale bar, 100 μm. hpf, hours post fertilization. (c) Immunofluorescence staining of mouse embryos at 108 hpf. Z-projection and single-section immunofluorescence images of non-target-, NANOG-, SMAD2-, HNF4A-, TCF12-, HBP1-, CDX2-, MED1- and ELF5-siRNA injected embryos, showing trophectoderm fate (CDX2, green), inner cell mass fate (SOX2, magenta), and DNA (DAPI, cyan). White arrowheads indicate the decrease in SOX2+ cells. Representative images out of three independent experiments are shown. Asterisks indicate the adjacent embryos. Scale bar, 100 μm. (d) UMAP showing individual embryos colored by specific knock-down (KD) TFs for 8 C embryos. The large colored dots indicate the median distribution of all embryos of each specific TF KD. (e-f) Violin plots showing the expression level of ICM signature genes (e), and TE signature genes (f) in 8-cell embryo RNA-seq data between non-targeting and TF KD groups. P value, two-sided Mann-Whitney test. Box plots center lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× IQR. The embryo number for each group in 8 C is as below: NANOG KD (20), ZFX KD (15), YY2 KD (26), TCF12 KD (19), CEBPB KD (19), BBX KD (14), SMAD2 KD (15), HBP1 KD (16), CDX2 KD (12), SOX15 KD (14), and NC (19). (g) UMAP showing individual embryos colored by specific KD TFs for blastocyst embryo. The large colored dots indicate the median distribution of all embryos of each specific TF KD. (h-i) Violin plots showing the expression level of ICM signature genes (h), and TE signature genes (i) in blastocyst embryo RNA-seq data between non-targeting and TF KD groups. P value, two-sided Mann-Whitney test. Box plots center lines indicate the median, box limits indicate the first and third quartiles, and whiskers indicate 1.5× IQR. The embryo number for each group in blastocyst is as below: HNF4A (19), KLF6 KD (10), ELF5 KD (20), HIF1A KD (13), and NC (15). (j-k) Smoothed heatmap showing the SCENIC TF activity score for different groups in 8-cell (j) and blastocyst embryos(k) RNA-seq data.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and legends for Supplementary Tables 1–10.
Supplementary Tables
Supplementary Tables 1–10.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, M., Yue, Y., Chen, X. et al. Genome-coverage single-cell histone modifications for embryo lineage tracing. Nature 640, 828–839 (2025). https://doi.org/10.1038/s41586-025-08656-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08656-1
This article is cited by
-
CRISPR-Based Functional Genomics in Pluripotent Stem Cells
Stem Cell Reviews and Reports (2026)
-
Folic acid ameliorates placental structure and function in fetal growth restriction via epigenetic modifications
Clinical Epigenetics (2025)
-
Slow awakening of the silent X chromosome in female primordial germ cells
Nature Structural & Molecular Biology (2025)
-
TACIT and CoTACIT for histone modification profiling in single cells and lineage tracing
Nature Reviews Genetics (2025)
-
Remodelling bivalent chromatin is essential for mouse peri-implantation embryogenesis
Nature Cell Biology (2025)