Abstract
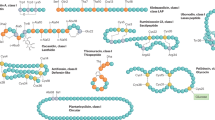
Lasso peptides (biologically active molecules with a distinct structurally constrained knotted fold) are natural products that belong to the class of ribosomally synthesized and post-translationally modified peptides1,2,3. Lasso peptides act on several bacterial targets4,5, but none have been reported to inhibit the ribosome, one of the main targets of antibiotics in the bacterial cell6,7. Here we report the identification and characterization of the lasso peptide antibiotic lariocidin and its internally cyclized derivative lariocidin B, produced by Paenibacillus sp. M2, which has broad-spectrum activity against a range of bacterial pathogens. We show that lariocidins inhibit bacterial growth by binding to the ribosome and interfering with protein synthesis. Structural, genetic and biochemical data show that lariocidins bind at a unique site in the small ribosomal subunit, where they interact with the 16S ribosomal RNA and aminoacyl-tRNA, inhibiting translocation and inducing miscoding. Lariocidin is unaffected by common resistance mechanisms, has a low propensity for generating spontaneous resistance, shows no toxicity to human cells, and has potent in vivo activity in a mouse model of Acinetobacter baumannii infection. Our identification of ribosome-targeting lasso peptides uncovers new routes towards the discovery of alternative protein-synthesis inhibitors and offers a novel chemical scaffold for the development of much-needed antibacterial drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting the findings of this study are available within this paper and its Supplementary Information or have been deposited to the indicated databases. Coordinates and structure factors were deposited in the RCSB Protein Data Bank (PDB) with the following accession codes: 9DFC for the wild-type T. thermophilus 70S ribosome in complex with LAR, mRNA, aminoacylated A-site Phe-tRNAPhe, aminoacylated P-site fMet-tRNAiMet, and deacylated E-site tRNAPhe; 9DFD for the wild-type T. thermophilus 70S ribosome in complex with LAR-B, mRNA, aminoacylated A-site Phe-tRNAPhe, aminoacylated P-site fMet-tRNAiMet, deacylated E-site tRNAPhe; and 9DFE for the wild-type T. thermophilus 70S ribosome in complex with LAR and protein Y. All previously published structures used in this work for structural comparisons were retrieved from the RCSB Protein Data Bank: PDB entries 6XHW, 6XHX, 6CAE, 4G5K, 4YBB and 4W2I. The complete genome sequence of Paenibacillus sp. M2 is available in NCBI GenBank under accession no. CP169648. Phylogenetic tree and the DNA segments containing lrc-like BGC are available at https://github.com/jangrm1/LAR-BGC. Source data are provided with this paper.
Change history
11 September 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41586-025-09597-5
References
Maksimov, M. O., Pan, S. J. & James Link, A. Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 29, 996–1006 (2012).
Tietz, J. I. et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478 (2017).
Barrett, S. E. & Mitchell, D. A. Advances in lasso peptide discovery, biosynthesis, and function. Trends Genet. 40, 950–968 (2024).
Gavrish, E. et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518 (2014).
Mukhopadhyay, J., Sineva, E., Knight, J., Levy, R. M. & Ebright, R. H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell. 14, 739–751 (2004).
Wilson, D. N. The A–Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44, 393–433 (2009).
Lin, J., Zhou, D., Steitz, T. A., Polikanov, Y. S. & Gagnon, M. G. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 87, 451–478 (2018).
Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Jesudason, T. WHO publishes updated list of bacterial priority pathogens. Lancet Microbe 5, 100940 (2024).
Liu, Y., Ding, S. Y., Shen, J. Z. & Zhu, K. Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat. Prod. Rep. 36, 573–592 (2019).
Sieber, S. A. & Marahiel, M. A. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005).
Ongpipattanakul, C. et al. Mechanism of action of ribosomally synthesized and post-translationally modified peptides. Chem. Rev. 122, 14722–14814 (2022).
Arnison, P. G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Pfeiffer, I. P., Schroder, M. P. & Mordhorst, S. Opportunities and challenges of RiPP-based therapeutics. Nat. Prod. Rep. 41, 990–1019 (2024).
Kuznedelov, K. et al. The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J. Mol. Biol. 412, 842–848 (2011).
Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585 (1995).
Smith, L. K. & Mankin, A. S. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52, 1703–1712 (2008).
Garneau-Tsodikova, S. & Labby, K. J. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7, 11–27 (2016).
Cook, M. A. et al. Lessons from assembling a microbial natural product and pre-fractionated extract library in an academic laboratory. J. Ind. Microbiol. Biotechnol. 50, kuad042 (2023).
Nation, R. L. & Li, J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535–543 (2009).
Hancock, R. E. & Rozek, A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206, 143–149 (2002).
Quan, S., Skovgaard, O., McLaughlin, R. E., Buurman, E. T. & Squires, C. L. Markerless Escherichia coli rrn deletion strains for genetic determination of ribosomal binding sites. G3 5, 2555–2557 (2015).
Orelle, C. et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 57, 5994–6004 (2013).
Polikanov, Y. S., Aleksashin, N. A., Beckert, B. & Wilson, D. N. The mechanisms of action of ribosome-targeting peptide antibiotics. Front. Mol. Biosci. 5, 48 (2018).
Pantel, L. et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell 70, 83–94.e87 (2018).
Peske, F., Savelsbergh, A., Katunin, V. I., Rodnina, M. V. & Wintermeyer, W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J. Mol. Biol. 343, 1183–1194 (2004).
Noller, H. F., Lancaster, L., Zhou, J. & Mohan, S. The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol. 24, 1021–1027 (2017).
Rundlet, E. J. et al. Structural basis of early translocation events on the ribosome. Nature 595, 741–745 (2021).
Orelle, C. et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 41, e144 (2013).
Olivier, N. B. et al. Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome. Proc. Natl Acad. Sci. USA 111, 16274–16279 (2014).
Brenner, S. & Beckwith, J. Ochre mutants, a new class of suppressible nonsense mutants. J. Mol. Biol. 13, 629–637 (1965).
Kohanski, M. A., Dwyer, D. J., Wierzbowski, J., Cottarel, G. & Collins, J. J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008).
Cao, L., Do, T. & Link, A. J. Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs). J. Ind. Microbiol. Biotechnol. 48, kuab005 (2021).
Carson, D. V., Juarez, R. J., Do, T., Yang, Z. J. & Link, A. J. Antimicrobial lasso peptide cloacaenodin utilizes a unique TonB-dependent transporter to access susceptible bacteria. ACS Chem. Biol. 19, 981–991 (2024).
Do, T., Thokkadam, A., Leach, R. & Link, A. J. Phenotype-guided comparative genomics identifies the complete transport pathway of the antimicrobial lasso peptide ubonodin in burkholderia. ACS Chem. Biol. 17, 2332–2343 (2022).
Miller, S., Goy, K., She, R., Spellberg, B. & Luna, B. Antimicrobial susceptibility testing performed in RPMI 1640 reveals azithromycin efficacy against carbapenem-resistant Acinetobacter baumannii and predicts in vivo outcomes in Galleria mellonella. Antimicrob. Agents Chemother. 67, e01320–e01322 (2023).
Luna, B. et al. A nutrient-limited screen unmasks rifabutin hyperactivity for extensively drug-resistant Acinetobacter baumannii. Nat. Microbiol. 5, 1134–1143 (2020).
Farha, M. A., French, S., Stokes, J. M. & Brown, E. D. Bicarbonate alters bacterial susceptibility to antibiotics by targeting the proton motive force. ACS Infect. Dis. 4, 382–390 (2018).
Andersen, F. D. et al. Triculamin: an unusual lasso peptide with potent antimycobacterial activity. J. Nat. Prod. 85, 1514–1521 (2022).
Ersoy, S. C. et al. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20, 173–181 (2017).
Polikanov, Y. S. et al. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol. Cell 56, 541–550 (2014).
Chen, Z., Erickson, D. L. & Meng, J. Benchmarking hybrid assembly approaches for genomic analyses of bacterial pathogens using Illumina and Oxford Nanopore sequencing. BMC Genomics 21, 631 (2020).
Blin, K. et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–w35 (2021).
Blin, K. et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–w50 (2023).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Gilchrist, C. L. M. & Chooi, Y.-H. clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475 (2021).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Bai, C. et al. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc. Natl Acad. Sci. USA 112, 12181–12186 (2015).
Hong, H. J., Hutchings, M. I., Hill, L. M. & Buttner, M. J. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280, 13055–13061 (2005).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Xu, M. et al. GPAHex-A synthetic biology platform for type IV–V glycopeptide antibiotic production and discovery. Nat. Commun. 11, 5232 (2020).
Cox, G. et al. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem. Biol. 24, 98–109 (2017).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Speers, A. E. & Cravatt, B. F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 11, 535–546 (2004).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Polikanov, Y. S., Blaha, G. M. & Steitz, T. A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 (2012).
Polikanov, Y. S., Steitz, T. A. & Innis, C. A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 21, 787–793 (2014).
Polikanov, Y. S., Melnikov, S. V., Soll, D. & Steitz, T. A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015).
Syroegin, E. A., Aleksandrova, E. V. & Polikanov, Y. S. Insights into the ribosome function from the structures of non-arrested ribosome–nascent chain complexes. Nat. Chem. 15, 143–153 (2023).
Aleksandrova, E. V. et al. Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it. Nat. Chem. Biol. 20, 867–876 (2024).
Svetlov, M. S. et al. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 17, 412–420 (2021).
Chen, C. W. et al. Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A. Nat. Commun. 14, 4196 (2023).
Mitcheltree, M. J. et al. A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature 599, 507–512 (2021).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Noeske, J. et al. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015).
Acknowledgements
The authors thank M. Cook for determining mycobacterial MICs; A. Guitor for help with breseq analysis of resistant mutants; M. Surette for MIC determination against gut microbiota; E. Brown for E. coli and B. subtilis gene-deletion strains; the McMaster Biointerfaces Institute for MALDI–MSMS analysis; the Centre for Microbial Chemical Biology for mammalian toxicity experiments; and the McMaster Centre for Advanced Light Microscopy facility for confocal microscopy. This work is based on research conducted at the Center for BioMolecular Structure beamlines (17ID-1 and 17ID-2), which are primarily supported by the National Institute of General Medical Sciences from the National Institutes of Health (P30-GM133893) and by the DOE Office of Biological and Environmental Research (KP1605010). NSLS2 is a US DOE Office of Science User Facility operated under contract no. DE-SC0012704. This publication resulted from the data collected using the beamtime obtained through NECAT BAG proposal 311950. This work was supported by the Canadian Institutes for Health Research (project grant PJT190298 to G.D.W.), National Institute of General Medical Sciences of the National Institutes of Health (grant R35-GM127134 to A.S.M.; grant R35-GM151957 to Y.S.P.; grant R01-GM132302 to Y.S.P.), National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant R01-AI162961 to A.S.M., N.V.-L. and Y.S.P.), National Science Foundation (MCB-2345351 to N.V.-L.) and the Illinois State startup funds (to Y.S.P.). The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
M.J., M.K. and G.D.W. conceived the study and planned initial experiments. M.K. isolated the soil strains and performed preliminary antimicrobial screening. M.J. isolated LAR and conducted initial chemical characterization. W.W. performed NMR experiments. M.J. and M.T. performed heterologous expression studies in Streptomyces. M.J. and K.K. did the chemical analysis and synthesized fluorophore conjugates. M.J., L.D. and B.K.C. designed animal studies, and L.D. performed animal experiments. B.K.C. supervised animal studies. M.K. and A.S. performed DNA preparation and whole-genome sequencing. M.J., D.Y.T., M.K. and D.K. performed the biochemical and microbiological experiments. E.V.A., D.Y.T. and Y.S.P. designed and performed X-ray crystallography experiments. M.J. and D.Y.T. performed bioinformatics analysis. A.S.M., N.V.-L., Y.S.P., X.C., Z.D, and M.T. developed and supplied Streptomyces strains for heterologous expression, G.D.W. designed and supervised the experiments. All authors interpreted the results. M.J., D.Y.T., A.S.M., N.V.-L., Y.S.P. and G.D.W. wrote and edited the manuscript with input from L.D. and K.K. All authors approved the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks A. Link and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Paenibacillus sp. M2 strain produces colistin and LAR.
(a) The antibacterial activity of partially fractionated extract from Paenibacillus M2 against A. baumannii C0286 (‘Ab’) and the following E. coli BW25113 strains: wild type (WT), colistin-resistant expressing mcr-1, and the antibiotic hypersusceptible ΔtolC/ΔbamB mutant (ΔT/ΔB). (b) Bioactivity assay of RP-HPLC fractions of the pre-fractionated (on SP-sepharose column) Paenibacillus sp. M2 extract against A. baumannii C0286 strain, showing the presence of two distinct antibiotics. (c) Liquid chromatography-mass-spectrometry analysis of fractions 7 and 21 (see panel b) shows the presence of LAR and colistin, respectively. The upper panel shows the extracted ion chromatogram, and the bottom panels represent mass spectra of corresponding fractions. LAR, lariocidin; LAR-B, lariocidin B, and LAR-C, lariocidin C.
Extended Data Fig. 2 LAR does not affect the bacterial cell envelope.
(a) Inner membrane permeabilization assay in E. coli TOP10 cells with pUC19 plasmid (containing the lacZ gene) using membrane-impermeable dye Ortho-nitrophenyl-β-D-galactopyranoside (ONPG). LAR (40 μg/ml) did not facilitate the uptake of ONPG, unlike colistin (5 μg/ml), which creates membrane pores. Data are representative of two independent experiments. (b) Membrane depolarization assay using DiOC2(3) dye. The fluorescence of the dye quenches when it enters depolarized cells (due to membrane potential disruption). LAR was used at 10xMIC (40 μg/ml). Protonophore, CCCP (20 μM) served as a positive control. Data represent three biological experiments, with error bars indicating SD of three replicates (c) Scanning electron microscopy images of E. coli treated with 10xMIC of LAR, showing no obvious changes in morphology or defects in the cell envelope. The images are representative of two independent samples.
Extended Data Fig. 3 Synthesis of lariocidin-fluorophore conjugates and confocal microscopy.
(a) Synthesis of lariocidin-fluorophore conjugates via click-chemistry. TEA, triethylamine; PyBOP benzotriazol-1-yloxytripyrrolidinophosphonium hexafluorophosphate; TFA, trifluoroacetic acid; DMSO, dimethylsulfoxide; DCM, dichloromethane. (b) MIC of LAR-fluorophores against E. coli BW25113 strain in MOPS minimal medium. (c, d) Confocal microscopy images of E. coli BW25113 cells treated with LAR-BODIPY (20 μg/ml) (c) or LAR-rhodamine (20 μg/ml) (d) indicating intracellular accumulation of probes. Fm-4-64 was used to stain the membrane, and Hoechst 33342 for DNA visualization. The images are representative of three biological replicates. Scale bar is 5 μM.
Extended Data Fig. 4 Lariocidin utilizes membrane potential to enter the bacterial cytoplasm.
E. coli BW25113 was used in all the experiments. (a) Anaerobic conditions result in an increase of LAR MIC determined in different media. MOPS MM is MOPS minimal medium and MHB is cation-adjusted Mueller-Hinton Broth. (b) Addition of bicarbonate, known to potentiate certain antibiotics like macrolides and aminoglycosides by enhancing the active membrane potential38, reduces LAR MIC in the MOPS and MHB media. MIC is also reduced in RPMI medium, which mimics physiological conditions better than MHB. (c) Effect of lower pH (known to decrease the membrane potential) on MIC. The experiment was conducted in MOPS MM. (d) The protonophore, CCCP, which eliminates the membrane potential, protects the cells from the killing action of LAR. cfu were enumerated 1 h after treatment with LAR (40 μg/ml) and/or CCCP (20 μM) in MOPS MM. However, no significant change in MIC of LAR was observed at this concentration of CCCP, suggesting that cells may overcome the effect of CCCP during prolonged growth. Data are plotted and mean ± SD of three biological replicates (e) LAR-BODIPY uptake under various treatment conditions using confocal microscopy showing that lowing pH or pretreatment of cells with the protonophore CCCP significantly reduces the accumulation of the LAR-BODIPY fluorescence inside the cells. Fm-4-64 was used to stain the membrane, and Hoechst 33342 for DNA visualization. The images are exemplary of two biological experiments. Scale bar is 5 μM.
Extended Data Fig. 5 LAR-resistance mutations in the 16S rRNA.
(a) Characteristics of LAR-resistant mutants selected in E. coli SQ110 ΔtolC. (b) Location of the mutations conferring increased resistance to LAR in the 16S rRNA helices h31 and h34. (c) Odilorhabdins (NOSO-95719) and LAR have different resistance profiles, as evidenced by the lack of LAR MIC changes in most NOSO-95719-resistant strains with point mutations in the 16S rRNA gene. (d) Spatial arrangement of the LAR-resistance mutations in the 16S rRNA shown in the structure of the E. coli small ribosomal subunit (PDB ID: 4YBB)69.
Extended Data Fig. 6 Intramolecular H-bonds of LAR and comparison of LAR and LAR-B structures.
(a, b) Structure of ribosome-bound LAR is shown from two opposite sides, highlighting intramolecular H-bonds that stabilize its fold. Carbon atoms are colored yellow, nitrogens are blue, oxygens are red. (c, d) Superposition of the structures of ribosome-bound LAR and LAR-B. Residues whose positions differ between the two isoforms are labeled. Nitrogen and oxygen atoms in the isopeptide bonds are colored blue and red, respectively.
Extended Data Fig. 7 LAR effectively kills A. baumannii C0286 in vitro and ex vivo.
(a) In vitro time-kill assay in MHB medium showing the bactericidal effect of LAR as denoted by reduction in viable cfu counts. Data are plotted as the mean of three biological replicates with the error bars indicating SD. (b) Cidality of LAR in the ex vivo model. A. baumannii C0286 was inoculated in human blood, and bacterial cfu were enumerated after 4 h treatment with LAR. Data are plotted as mean ± SD of three biological experiments. Significance was determined using one-way ordinary ANOVA with Dunnett’s multiple comparisons test. (*P = 0.0118; **P = 0.0017). (c) MIC of LAR against A. baumannii C0286 in MHB with or without the addition of serum. FBS=fetal bovine serum; HS=human serum (heat inactivated).
Extended Data Fig. 8 lrc-like biosynthetic gene clusters in various bacterial genomes.
(a) Gene composition of the representative set of lrc-like BGCs from different bacterial phyla. Each gene is represented by an arrow, and the proposed functions of the encoded proteins are listed on the right. The lrc BGC from Paenibacillus sp. M2 is labeled in bold. Note that BGCs from Actinomycetota (including the BGC of triculamin from S. griseocarneus ATCC 29818 (ref. 35)) lack the homologs of lrcB1B2 genes as the encoded precursors contain the unusual C-terminal follower peptide instead of the N-terminal leader peptide typical for other LPs. (b) Phylogenetic tree of lrc-like BGCs (n = 29) built based on the amino acid sequence similarity between LrcC (lasso cyclase) homologs. The lasso cyclase from the biosynthetic gene cluster of the lasso peptide paeninodin served as the outgroup. The alignment of the amino acid sequences of the precursor peptide(s) core parts is shown on the right. The amino acids are colored according to their physico-chemical properties. The consensus sequence and the extent of sequence conservation for each position are shown below. Note that for three triculamin-like peptides from Actinomycetota containing a follower rather than the leader peptide, the actual cleavage site for the follower-peptide (marked with black asterisks) could not be identified unambiguously. RRE – RiPP recognition element, GNAT – GCN5-Related N-acetyltransferase, OM – outer membrane.
Extended Data Fig. 9 Common mechanisms conferring resistance to clinically relevant ribosome-targeting antibiotics do not impact LAR antibacterial activity.
The graph shows the MIC increase of LAR and corresponding control antibiotics (Ab) upon overexpression of the designated resistance determinants in E. coli BW25113 ΔtolCΔbamB. The strain design and details of plasmids are described by Cox et al.52 The color of the gene name reflects the mechanism of resistance: antibiotic modification/inactivation – black, rRNA modification – orange, ribosome protection – blue. Functions of specific genes are- aad(3′′)(9)= spectinomycin adenyltransferase; apmA, aac(2′)-Ia, aac(6′)-Ib, and aac(2′)-IIa= aminoglycoside N-acetyltransferases; aph(4)-Ia, aph(3′)-IIIa, aph(3′)-IVa, and aph(6)-Ia= aminoglycoside phosphotransferases; sat= streptothricin acetyltransferase; tetX= tetracycline inactivation; vph= viomycin phosphotransferase; cat= chloramphenicol acetyltransferase; linB= lincosamide nucleotidyltransferase; vatD= streptogramins acetyltransferase; vgb= streptogramin B lyase; kamB, npmA, armA and rmtB = 16S rRNA methyltransferases; cfrA and ermC = 23S rRNA methyltransferases. Control antibiotics (Ab)- apramycin (apmA, kamB, npmA), gentamicin (aac(2′)-IIa, armA, rmtB), hygromycin B (aph(4)-Ia), kanamycin (aac(6′)-Ib, aph(3′)-IIIa), kasugamycin (aac(2′)-IIa), ribostamycin (aph(3′)-IVa), streptomycin (aph(6)-Ia), clindamycin (cfrA, linB, ermC), flopristin (vatD), quinupristin (vgb), nourseothricin (sat), oxytetracycline (tetM, tetX), viomycin (vph). For the controls marked with asterisks (*), the actual MIC change value is higher than that of the one presented; this reflects the growth of bacteria in the presence of the antibiotic in the highest concentration tested. Antibiotics targeting the small ribosomal subunit are in the light blue background, large ribosomal subunit – is in the light orange.
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–14 and Tables 1–6. These figures and tables are for the chemical and biological characterization of LAR(s), plasmid pIJ10256-lrc, details of spontaneous resistant mutants, 70S ribosome crystal structure with LAR and protein Y, raw blots and list of strains, plasmids and primers used.
Supplementary Video 1
Illustration of LAR binding to the bacterial ribosome. The video consecutively shows: (1) zoom-out and (2) close-up views of the LAR-binding site adjacent to the A site on the small subunit of the T. thermophilus 70S ribosome; (3) lasso-like structure of the LAR peptide; (4) 2Fo − Fc Fourier electron density map of ribosome-bound LAR (blue mesh); and (5) details of LAR interactions with the 16S rRNA and the A-site tRNA. The E. coli numbering of the 16S rRNA nucleotides is used. Hydrogen bonds between LAR, rRNA and A-site tRNA are indicated with dashed lines.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jangra, M., Travin, D.Y., Aleksandrova, E.V. et al. A broad-spectrum lasso peptide antibiotic targeting the bacterial ribosome. Nature 640, 1022–1030 (2025). https://doi.org/10.1038/s41586-025-08723-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08723-7
This article is cited by
-
Repurposing lusutrombopag: unveiling anti-Staphylococcus aureus activity in a novel oral thrombopoietin receptor agonist
BMC Microbiology (2026)
-
Antimicrobial effects of Bacillus safensis BS05 cell free supernatant against Campylobacter jejuni and its probiotic evaluation
BMC Microbiology (2026)
-
Comprehensive genotypic report of carbapenem-resistant Klebsiella pneumoniae (CRKP) harboring carbapenemase-resistant genes, porin loss and efflux pump genes in Osun State, Nigeria
Molecular Biology Reports (2026)
-
The application of AI-driven and engineered intratumoral microbes in cancer therapy
Journal of Translational Medicine (2025)
-
Lasso-shaped molecule is a new type of broad-spectrum antibiotic
Nature (2025)