Abstract
Long (>200 nucleotides) non-coding RNAs (lncRNAs) play important roles in diverse aspects of life. Over 20 classes of lncRNAs have been identified in bacteria and bacteriophages through comparative genomics analyses, but their biological functions remain largely unexplored1,2,3. Owing to the large sizes, the structural determinants of most lncRNAs also remain uncharacterized. Here, we report the structures of two natural RNA nanocages formed by the ROOL (rumen-originating, ornate, large) lncRNA found in bacterial and phage genomes. The cryo-electron microscopy (cryo-EM) structures at 2.9-Å resolution reveal that ROOL RNAs form an octameric nanocage with a diameter of 28 nm and an axial length of 20 nm, in which the hollow inside features poorly ordered regions. The octamer is stabilized by numerous tertiary and quaternary interactions, including triple-strand A-minors, for which we propose the term ‘A-minor staples’. The structure of an isolated ROOL monomer at 3.2-Å resolution indicates that nanocage assembly involves a strand-swapping mechanism resulting in quaternary kissing loops. Finally, we show that ROOL RNA fused to an RNA aptamer, transfer RNA or microRNA retains its structure, forming a nanocage with radially displayed cargoes. Our findings, therefore, may enable engineering of novel RNA nanocages as delivery vehicles for research and therapeutic applications.
Similar content being viewed by others
Main
Highly structured non-coding RNAs (ncRNAs) contribute to the most fundamental biological processes. Well-known examples include ribosomal and transfer RNAs (rRNAs and tRNAs) that perform protein synthesis, group II introns mobilizing genetic elements4 and signal-recognition particle RNAs that target secretory proteins to the membrane5. Comparative genomics analyses have identified hundreds of other structured ncRNAs in bacteria6,7, including approximately 20 classes of long ncRNAs (lncRNAs) whose functions remain unknown1,2,3. ROOL (rumen-originating, ornate, large) RNAs of approximately 600 nucleotides, originally discovered in metagenomics data from cow rumen, have been identified in bacteriophage genomes and at least three phyla of bacteria2, many of which are opportunistic pathogens that inhabit the human mucosa and contribute to diseases (Fig. 1a). An even larger, approximately 800-nucleotide GOLLD (giant, ornate, lake- and Lactobacillales-derived) lncRNA is smaller only than 23S and 16S rRNAs and proposed to be involved in bacteriophage lysis, but its exact role is still unknown1. Both ROOL and GOLLD RNAs often reside next to tRNAs (sometimes overlapping by a few nucleotides) in the genome and can be found in prophages, suggesting that they may have related biological functions2. ROOL RNA has unusually high expression, comparable to that of 16S rRNA, in some strains of Lactobacillus salivarius, although its deletion from L. salivarius caused no obvious phenotype under laboratory conditions8.
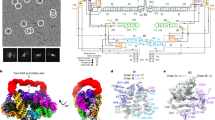
a, ROOL-encoding bacteria are associated with diseases in humans. Circle sizes and colours indicate enrichment scores of each bacteria species or group for certain diseases available from the Human Gut Microbiome Atlas. The two ROOL RNA-bearing bacteria species or groups used in this study and their potentially associated human diseases are labelled in red. b,c, Cryo-EM maps of ROOLEfa (σ = 2.88) (b) and ROOLFirm (σ = 4.16) (c) determined at 2.9-Å resolution; top, side and bottom views are shown, with ROOL monomers individually coloured. d, Alignment of ROOLEfa (σ = 3.88) and ROOLFirm (σ = 3.97) maps demonstrates similar architectures. ROOLEfa and ROOLFirm are blue and brown, respectively.
Here, we used single-particle cryo-electron microscopy (cryo-EM) analysis to determine the structures of two 580-nucleotide long ROOL RNAs: ROOLEfa from Enterococcus faecalis JH1 (2.94 Å) and ROOLFirm from Firmicutes bacterium CAG:227 (2.93 Å), both of which are Gram-positive bacteria that belong to the Bacillota (Firmicutes) phylum. Enterococcus faecalis JH1 is an isolated clinical strain that can contribute to bacteraemia, endocarditis and urinary infection, whereas Firmicutes bacterium CAG:227 belongs to the Lachnospiraceae family, which is among the most abundant taxa in the human gut microbiota. We find that these ROOL RNAs form highly similar cage-like structures composed of eight ROOL monomers. In addition, we obtained the individual ROOLEfa monomer structure at 3.2-Å resolution. Comparison of the monomer with the octamer suggests an assembly mechanism involving strand swapping. Furthermore, we explored the possibility of using ROOL RNA as a nanocarrier. Fusing various RNA cargoes at the 3′ end of a ROOL RNA does not disrupt the octameric structure, highlighting the potential of using ROOL to stabilize and deliver RNA or other cargoes for research and therapeutics.
Architecture of ROOL octamers
Both ROOLEfa and ROOLFirm form octameric cage-like structures with a 28-nm diameter and 20-nm axial length (Fig. 1b–d and Extended Data Figs. 1–4). The octamers consist of top and bottom halves, each containing four monomers (Fig. 1b,c). Each monomer consists of 16 helical regions (H1–H16) forming extensive tertiary interactions to stabilize the slightly curved shape (Fig. 2a,h and Extended Data Fig. 5a,h). These include kissing loops, triple-strand A-minor interactions, base stacking, Z-anchors and base triple-mediated 90° turns (Fig. 2, Extended Data Fig. 5 and Supplementary Video 1). We propose the term ‘A-minor staples’ to describe the triple-strand A-minor interactions, in keeping with the nomenclature of three-strand junctions9. Kissing loops and A-minor interactions form the majority of interhelical interactions. In ROOLEfa, the most prominent are the kissing loops of H11 and H12 at nucleotides 303–306 and 359–362, held together by four Watson–Crick base pairs (Fig. 2d). As discussed below, this interaction appears to be crucial for the octamer assembly. The A-rich loop of H6 also engages in a kissing loop-like interaction with the A-rich internal loop of H8 (Fig. 2e), as the adenosines 148 and 149 form base triples with the U205–A336 and C204–G337 pairs. In addition, ROOLEfa features numerous intramolecular A-minor interactions, the most abundant tertiary interaction in most RNAs9. A-minor interactions usually involve two consecutive adenosines packed against a minor groove of an RNA helix, so that the riboses are placed next to each other to hydrogen bond. They play crucial roles in RNA stabilization and molecular recognition, including tetraloop–receptor recognition10,11,12,13, mRNA decoding by tRNA14,15,16 and transfer-messenger RNA (tmRNA) stabilization within the ribosome17. In ROOLEfa, an intriguing A-minor staple involving A218 and A219, with A96 stacked on top, holds together three helical elements: H4, H9 and H10 (Fig. 2f). This junction is buttressed by another A-minor staple involving stacked adenosines A295, A296 (H10) and A216 (H9) docked at H4 (Fig. 2g). In both A-minor staples, the non-consecutive adenosine provides the stacking platform for the consecutive adenosines, but its ribose does not form hydrogen bonds with the corresponding minor groove. As discussed below, these interactions also enable the monomer to adopt the octamer-compatible conformation.
a, Molecular model of a ROOLEfa monomer (Mono1) within the octameric structure. b–g, Close-up views of examples of tertiary interactions stabilizing the monomer are shown in the corresponding coloured boxes with the map rendered as a transparent surface (σ = 2.88): a Z-anchor (b), a 90° turn (c), the H12–H11 kissing loop (d), an H6–H8 interaction (e), an A-minor staple joining helices H4, H9, and H10 (f), a second A-minor staple involving these helices (g). h, Secondary structure annotation of ROOLEfa within the octamer, with tertiary interactions labelled by coloured boxes that match panels b–g. Nucleotides in panels b–h are coloured according to helix colours in a.
To probe the oligomerization states of ROOL RNAs in solution, we characterized ROOLEfa and ROOLFirm by mass photometry and size-exclusion chromatography (SEC). At a concentration of 50–100 nM, monomer and octamer were the two major molecular species detected by mass photometry. Nevertheless, dimer, tetramer, decamer and octadecamer species were also detected, suggesting that ROOL can form additional types of oligomers (Extended Data Fig. 1b). Consistent with these results, octamers and monomers eluted in two major peaks from an SEC column (Extended Data Fig. 1c–e). Negative-stain electron microscopy and dynamic light scattering data further corroborate the SEC and mass photometry data (Extended Data Fig. 1f,g), underscoring the predominance of octameric and monomeric species discussed below.
Intermolecular interactions in ROOLEfa
The radial array of ROOLEfa monomers that form each tetramer is stabilized primarily by kissing-loop and A-minor interactions (Fig. 3a–f and Extended Data Fig. 6). The four-fold axis is surrounded by the H9 helices of each monomer docked into each other. Here, the loop nucleotides G231 and U232 of one monomer pair with the intrahelical C224 and A223 of a neighbouring monomer (Fig. 3a–c). Each tetramer contains four dimeric interfaces, in each of which the monomers are held together by four major interactions: the H9 docking described above, two kissing-loop contacts and an A-minor interaction (Fig. 3b–f). The H10 loop (at nucleotides 262–264) forms a kissing-loop interaction with the internal loop between H8 and H10 (at nucleotides 287–289), featuring three Watson–Crick pairs (Fig. 3d). Notably, this interaction is possible because the internal loop curves around H4, held in place by the A-minor staple interactions described above (Fig. 2f,g). The other kissing-loop contact is formed by the loop of H2 of one monomer protruding into H12 of the second monomer to form four Watson–Crick pairs between nucleotides 42–45 and 420–423, respectively (Fig. 3e). This stem is coaxially stacked on the rest of H12, whose backbone forms a ribose zipper with the intramolecular H11–H12 kissing loop described above (Fig. 2d). This underscores the intricate dependence of the higher order packing on the internal monomer conformation, implied in the oligomerization described below. The A-minor interaction involving three consecutive adenosines A318–320 brings together H11 of one monomer together with the sharply curved H3 of the neighbouring monomer (Fig. 3f). Here, A318 stacks on the adenosine of the Hoogsteen–ribose A166–G65 pair, whereas A319 and A320 pack against the minor groove formed by G64 and G167–U63, respectively (Fig. 3f).
a, Top front view of the ROOLEfa tetramer looking down the four-fold axis. Close-up views of two key interactions are shown in panels c and d with map rendered as a transparent surface (σ = 2.88). b, The ROOLEfa dimer is stabilized by four key interactions. c–f, Close-up views of the four key interactions shown in b, with map rendered as a transparent surface (σ = 2.88): base pairing between H9 helices (c), the kissing loop between H10 helices (d), the kissing loop between H2 and H12 (e), A-minor interactions between H3 and H11 (f). The inside view in panel b was obtained by flipping the molecule in panel a (top view). Coloured boxes in panels c and d show close-up views of those in panels a and b; residues are coloured to match panel b. g, Molecular model of a ROOLFirm monomer within the octameric structure. h, The alignment of ROOLEfa and ROOLFirm structures of monomers within the corresponding octamers shows highly similar architecture. ROOLEfa and ROOLFirm are coloured blue and brown, respectively.
The interactions between tetramers (to form an octamer) are less extensive than those between monomers within a tetramer. The interface between the monomers from two interacting tetramers is symmetrical, including three pairs of contacts (Extended Data Fig. 7i–k). First, the base of G443 at the tip of H13 is docked into H7 of the second monomer, sandwiched between A187 and A535 and forming a Hoogsteen pair with C186 (Extended Data Fig. 7j). Next, A502 from the internal loop of H15 packs onto the ribose of A555 from the corresponding ROOLEfa partner (Extended Data Fig. 7k). Finally, at the periphery of the dimeric interface, two hairpin loops of H15 approach each other, but the low map resolution in this region precludes detailed modelling and indicates a weak contact.
Analyses of ROOLFirm and its variants
To corroborate our findings of the octameric ROOLEfa, we determined the structure of ROOLFirm, a ROOL family RNA from a Firmicutes bacterium. Despite substantially different primary sequences (Fig. 2h, Extended Data Fig. 5h and Supplementary Table 1), the structure of ROOLFirm is almost identical to that of ROOLEfa (Figs. 1d and 3h and Extended Data Figs. 5–7), highlighting a conservation of RNA quaternary structure. One notable difference is that ROOLFirm tetramers interact with each other via G-A base stacking, whereas ROOLEfa tetramers interact via Hoogsteen G•C base pairing (Extended Data Fig. 7g,j). Many differences in intratetramer interactions include distinct base-pairing or kissing-loop compositions; however, their positions and secondary structure patterns in ROOLFirm and ROOLEfa are alike. The high similarity of intratetramer interactions and the ability of ROOL octamers to adopt slight changes at the intertetramer interface underscore the importance of the conserved octameric quaternary structure for the cellular function(s) of ROOL RNAs.
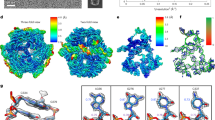
Three-dimensional (3D) alignments showed that ROOLFirm and ROOLEfa octamers do not perfectly overlap owing to a slight register shift in the tetrameric packing (Figs. 1d and 3h). We noted that the shape of the octamers might be affected by the disordered linker regions within the nanocage (nucleotides 365–420 in ROOLEfa, nucleotides 67–96 and 385–413 in ROOLFirm) that form low-resolution density insufficient for detailed structural modelling (Fig. 4a). To test whether these regions are critical for octamer assembly, we generated ROOLFirm mutants lacking one or both of these regions (Fig. 4b). Negative-stain electron microscopy showed that the shapes of the particles are nearly identical to that of the wild-type ROOLFirm, further emphasizing that the octamers are held primarily by the tertiary and quaternary interactions resolved in our cryo-EM maps (Fig. 4c).
a, Poorly resolved (disordered) regions are located in the cavities of ROOLFirm and ROOLEfa nanocages. These regions (D1, D2 in ROOLFirm and D, D′ in ROOLEfa) are coloured in low pass-filtered maps. The Gaussian filter with width 1.64 for the sharpened cryo-EM map is displayed at the following contours: 2.74σ for ROOLFirm, 2.49σ for ROOLEfa. b, Diagrams of ROOLFirm wild type (WT) and deletion mutants tested in this work, in which the disordered regions were removed. Each coloured box represents a helix scaled to length in the structure. c, Negative-stain analysis indicates that ROOLFirm deletion mutants without disordered regions can form nanocages similar to those formed by WT ROOLFirm. This experiment was performed once with two micrographs collected for each RNA. d, Design of ROOL–cargo RNA fusions. e, When fused to a Mango-III aptamer, pre-tRNA or primary microRNA (pri-miRNA) at the 3′ end, ROOLFirm forms stable nanocages with radially displayed cargoes demonstrated by negative-stain electron microscopy (micrographs shown) and corresponding 2D averages (examples of 12 classes are shown for each construct). This experiment was performed once with >70 micrographs collected for each RNA. For original and additional negative-stain micrographs, see Supplementary Fig. 1 and data deposited to Figshare35. Scale bars, 50 nm (c), 100 nm (e).
Conserved extensive interactions within the tetramers of both ROOLFirm and ROOLEfa suggest that tetramers could form assembly intermediates of the octamer. To test whether isolated tetramers exist in our datasets, we performed data processing using a low pass-filtered tetramer map (20 Å) derived from the ROOLFirm or ROOLEfa octamers. In the ROOLFirm dataset, a small fraction of tetramer particles (approximately 8% of the octamer particles) resulted in a higher resolution (3.59 Å) reconstruction featuring a tetramer conformation nearly identical to that within the octamer (Extended Data Fig. 3). The ROOLEfa dataset did not yield a substantially improved reconstruction, probably as a result of an insufficient dataset size, which is approximately half that of ROOLFirm (Extended Data Table 1). Nevertheless, this analysis indicates that tetramers can form independently of octamers, which was also observed by mass photometry analysis (Extended Data Fig. 1b), suggesting that they may act as assembly and/or disassembly intermediates.
In both ROOL octamers, the 5′ and 3′ termini form H1 protruding away from the nanocage (Fig. 3a). We asked whether the termini of ROOL can be fused with another RNA without disrupting the nanocage, to test its potential utility as a nanocarrier for RNAs or other molecules. To this end, we created three constructs in which either of the three RNA cargoes were fused to the 3′ end of ROOLFirm via in vitro transcription. These included the 51-nucleotide Mango-III aptamer18,19, which can be used for imaging ROOL RNA using fluorescence, a 163-nucleotide suppressor tRNA precursor and a 143-nucleotide primary microRNA containing human miR-1-1 (Fig. 4d). The suppressor tRNA comprises tRNASer modified to achieve high stop-codon readthrough activity in human cells and mice, to potentially be used in therapeutics20. Negative-stain electron microscopy demonstrates that all three constructs retain the shape consistent with the octamers observed with the wild-type ROOL (Fig. 4e). Furthermore, additional densities radiating from ROOLFirm particles in two-dimensional (2D) classes indicate relatively ordered densities, probably restrained by the stiff helix H1 (Fig. 4e). In sum, perturbations of both the intracage sequences and the outer-end termini, including long 163-nucleotide fusions, do not disrupt ROOLFirm multimerization.
Structure of ROOLEfa monomer
Our 2D and 3D classifications of ROOLEfa cryo-EM datasets revealed the presence of unassembled ROOLEfa monomers, whose structure was resolved to 3.25 Å (Fig. 5a and Supplementary Video 2). The core shape of the isolated monomer is similar to that within the octameric nanocage (Fig. 5b), but two major differences suggest large restructuring of the helical regions involved in octamer formation. The first notable rearrangement involves helices 8, 9 and 10. In the octameric monomer, these helices pack against H4, via A-minor interactions, to expose the internal loop between H8 and H10 (at nucleotides 286–292) for the intermolecular kissing-loop interaction (Fig. 3b,d). In the monomer, however, H8–H9 is shifted away from H4 by approximately 23 Å, and the internal loop appears to form a continuation of H8, whereas helices 9 and 10 are disordered (Fig. 5a,c,e). This region of the monomer, therefore, features a restructured strand required for intermolecular interactions in the octamer (Fig. 5c,e and Supplementary Video 2). The second notable difference involves H12. Instead of the compact intramolecular kissing-loop interaction (at nucleotides 359–362; Fig. 2d) connected by the poorly ordered linker at nucleotides 365–420 within the octamer, the monomer features a >100-Å long helix comprising residues 346–433 of H12 (Fig. 5a,b). Although lower resolution density does not enable modelling of the whole helix, the stem of the helix is clearly visible near the core of ROOLEfa RNA (Fig. 5a). Here, residues 420–423, which in the octamer are exposed to form intermolecular base pairs (Fig. 3e), are paired within the helix and stabilized by intrahelical stacking interactions (Fig. 5d). Strand swapping, involving approximately 41 Å of movement, is therefore required in this region to engage in intermolecular interactions with the loop of H2 (Figs. 2d, 3e and 5c,d and Supplementary Video 2).
a, Cryo-EM map of ROOLEfa monomer at 3.25 Å resolution (σ = 12.02). b, Comparison of ROOLEfa individual monomer with the monomer within the octameric assembly. c–e, Comparison of the individual monomer with the dimer within the octamer (c) details the rearrangements required to form two kissing-loop interactions (d,e). d, Close-up view of H12 in the monomeric and octameric conformations. e, Close-up view of H9 and H10 in the monomeric and octameric conformations. The individual monomer in b–e is coloured dark blue, the monomer within an octamer is coloured in green in b, in helix colour in c–e and the neighbouring monomer within an octamer is coloured in light blue in c–e.
To test the role of strand swapping in nanocage formation, we designed mutants that strengthen the pairing within the H12 helix in the monomer to disfavour new interactions with H2 and H11 (Extended Data Fig. 8a). Introducing a G opposite to the bulged C423, or changing the bulged AAU (417–419) to a C to enable pairing with G360, decreases nanocage formation as assayed by negative-stain electron microscopy, SEC and mass photometry (Extended Data Fig. 8). The combination of these two mutations has an even stronger effect, consistent with the notion that flexibility within H12 enables the strand swapping required for octamer assembly. By contrast, mutants that strengthen the pairing downstream of the regions involved in the kissing-loop formation are much less disruptive to nanocage formation (Extended Data Fig. 8). Echoing our findings for the disordered regions in ROOLFirm being disposable for nanocage assembly, these results outline possible engineering avenues to control cargo binding and octamer assembly.
We next tested the effect of salt concentration on the monomer–octamer equilibrium. Consistent with the stabilization of RNA tertiary structure by monovalent and divalent cations21,22, negative-stain electron microscopy, SEC and mass photometry demonstrate higher abundance of the octamer at 240 mM K+ and 20 mM Mg2+ (Extended Data Fig. 9). At 100 mM K+ and 10 mM Mg2+, which are closer to physiological concentrations in bacteria23, the equilibrium is shifted towards the monomer (Extended Data Fig. 9). The sensitivity of the monomer–octamer equilibrium to buffer conditions suggests that oligomer assembly in cells is regulated by ions and/or additional molecules.
Discussion
Structural implications of ROOL octamers
Bacteria harbour many types of organelles, which serve diverse functions, including storage or sequestration of specific metabolites and enzymes, enhancing spatial control of biological processes and increasing metabolic efficiency24. The striking nanocage structure of ROOL RNAs described here hints at their function as an encapsulation or transport vehicle for bacteria or bacteriophage. Its cavity is big enough to accommodate tRNA (with or without EF-Tu, the most abundant tRNA-binding protein), mRNA or other RNA/ribonucleoprotein complexes, but it is not spacious enough to host ribosomes or ribosomal subunits. Ribosomes were reported to crosslink with the much larger GOLLD RNA25, but it remains to be studied whether ribosomes or components of the translational machinery can be packed inside. When our work was finalized, Kretsch et al. reported ROOL and GOLLD nanocage structures26. In addition to showing that multimers of the approximately 800-nucleotide GOLLD RNA form large nanocages, the authors report that ROOL env-120, despite being a longer 659-nucleotide RNA, forms an octameric nanocage similar to that formed by the ROOLEfa and ROOLFirm RNAs. The overall similar architectures and diameters of nanocages formed by ROOL RNAs with different primary sequences and lengths points at a conserved function that remains to be uncovered.
The structures of ROOL RNAs revealed extensive intra- and intermolecular interactions that offer insights into the functional assembly and disassembly of the multimeric nanocage. Prevalent kissing-loop and A-minor interactions, including A-minor staples, underlie the intramolecular structure of the large ROOL molecules, as well as their intermolecular packing into tetramers. These unique interactions probably mediate the dynamic regulation of ROOL nanocages in cells. For example, disruption of the intertetramer interactions, which appear weaker than those holding the tetramers together, could allow the tetramers to separate, opening the nanocage to load or unload cargo. By contrast, disruption of the stronger intratetramer interactions, perhaps regulated by other cellular molecules, would probably result in the disassembly of the nanocage to release the cargo or to downregulate its function. Although the poorly ordered regions within the cage do not appear to substantially affect octamer assembly, they may play functional roles, such as the binding of specific cargoes, which remain to be uncovered.
Our work provides insights into the interactions critical for octamer assembly. Comparison of the ROOL octamer with the isolated monomer structure reveals that only two local strand swaps could enable two kissing-loop interactions to mediate the assembly of monomers into dimers. The RNA strand-swapping mechanism bears a notable resemblance to protein strand swapping driving oligomerization27,28. This similarity emphasizes that comparable strategies for increasing the architectural complexity have evolved for these fundamentally different biological polymers. The location of the intermolecular contacts within the ROOL dimer suggests the possibility of co-transcriptional assembly. The 5′-most region involved in the intradimer interactions comprises H4, which is well ordered in the monomer and forms the foundation for the intramolecular packing of helices 8 to 10. It is possible, therefore, that interactions with another monomer at this region—sampling the kissing-loop contact at the internal loop between H8 and H10 (nucleotides 286–292)—could form first during ROOL transcription. The second key region is the long helix at nucleotides 346–433 comprising H12. Consistent with its dynamic behaviour (Fig. 5a), this helix may transiently sample other conformations and engage in a dimeric interaction seeded by the previously formed contact via H4 and H8–10. Formation of all four key contacts, including the two kissing loops and an A-minor, would stabilize the dimer, forming the foundation for further oligomerization into the tetramer then octamer. Indeed, the same kissing-loop and A-minor interactions join the dimers into the tetramers observed in our data, which can ‘click’ into each other to form the octamer via symmetrical base stacking and base–ribose packing interactions.
RNA nanocages as delivery vehicles
Identification of the intricate tertiary and quaternary interactions in the natural RNA nanocage may help improve structure determination and design by deep learning29 and guide the emerging development of nanoparticles as drug delivery vehicles30. RNA nanostructures, including nanocages and origamis offer great thermodynamic stability along with pharmacokinetic and pharmacodynamic profiles31. Indeed, unlike most administered nucleic acids, which accumulate and clear in the liver, RNA nanocages may have different tissue distribution, including predominant accumulation in tumours32. Furthermore, RNA nanostructures can be used as modular vehicles to deliver RNA therapeutics, such as small interfering RNA precursors, for maturation and efficient gene silencing in human cells33. The ability of RNA oligomers to carry several copies of a therapeutic RNA can correlate with increased functional output, such as gene silencing33. The rapidly evolving space of RNA nanocages, however, primarily relies on designs involving base pairing and a subset of simple tertiary interactions31. Natural RNA nanocages could help expand therapeutic applications by advancing the thermodynamic, pharmacological and therapeutic properties of polymeric RNAs. For example, modifications of kissing-loop base pairing in ROOL octamers may modulate the stability of the octamer during and after delivery to cells. Future nanoparticle designs to improve the stability and solubility of RNA nanocages, facilitate cellular entry and intracellular trafficking and enable targeted delivery may therefore overcome the limitations of existing RNA oligomers and conventional drug delivery systems30. The large size and modularity of ROOL particles may render them novel nanocarriers with high stability, distinct cellular and tissue distributions and minimal immunogenicity34. Our study shows that removing the poorly ordered loops within the ROOL cage does not visibly perturb the octamer, paving the way for designing intracage cargoes. Furthermore, an aptamer or precursors of tRNA or microRNA can be fused at the ROOL termini without destabilizing the octameric structure. We anticipate that, as a natural RNA nanostructure, ROOL can be engineered in many ways for therapeutic development.
Methods
RNA preparation
The ROOLEfa and ROOLFirm sequences were derived from Rfam (accession numbers URS0000D65D6E_565648 and FR891750.1). The DNA templates for RNA synthesis were cloned into plasmids and amplified (gene and primer sequences are provided in the Supplementary Table 1). In vitro transcription of all RNAs was carried out with purified recombinant T7 RNA polymerase, 0.3 µM DNA template, 5 mM NTPs and 1 U µl−1 RNase inhibitor in 1× transcription buffer containing 50 mM Tris–HCl (pH 7.9), 0.01% TritonX-100, 20 mM MgCl2, 2 mM spermidine and 10 mM DTT at 37 °C for 3 h. The transcription products were mixed with 2× denaturing polyacrylamide gel-loading buffer containing 95% formamide, 0.025% SDS, 10 mM EDTA, 0.025% xylene cyanol and 0.025% bromophenol blue, and loaded on an 8 M urea, 4% polyacrylamide (19:1 acrylamide:bis) gel. The gel was run at 15 W for 3 h and visualized briefly using a 254-nm UV lamp held far from the gel to minimize RNA damage. Then, the RNA was eluted from the gel overnight in elution buffer containing 20 mM Tris–HCl (pH 7.5) and 300 mM NaCl on an active rotator at 4 °C. The resulting gel slurry was then filtered through 0.45-μm filters, RNA was precipitated by adding an equal volume of isopropanol and RNA pellets were washed two or three times with 75% ethanol and air dried. Precipitated RNA was resuspended in RNase-free water. The RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and stored at −80 °C. Polyacrylamide gel analyses of RNA samples are shown in Extended Data Fig. 1.
RNA refolding
For all subsequent experiments, RNAs were refolded at 0.125 μM in a refolding buffer containing 40 mM HEPES (pH 7.5) and 240 mM KCl or 100 mM KCl. The RNAs were heated from 23 °C to 90 °C, incubated for 3 min and cooled at 0.5 °C s−1 to 60 °C, MgCl2 was added to a final concentration of 20 mM or 10 mM or 2 mM and then samples were cooled to 25 °C for 30 min, incubated at 37 °C for 2 h and cooled to 4 °C before use. For cryo-EM, the sample was prepared in a buffer containing 40 mM HEPES (pH 7.5), 240 mM KCl and 20 mM MgCl2.
Size-exclusion chromatography analysis
RNA was folded as described above. A 50-µl refolded sample was separated on an S6 increase 3.2/300 column (Cytiva) with a total volume of 2.4 ml using a flow rate of 0.04 ml min−1 on an ÄKTA pure micro (Cytiva). Fractions of 0.02 ml were collected, and the peak fractions were analysed by 6% denaturing PAGE and negative-stain electron microscopy. Each sample was analysed twice.
Mass photometry analysis
ROOL RNA samples were folded as described above, and the Millennium RNA Marker (Thermo Fisher Scientific) was used for calibration. Samples were applied to a Superior Precision Cover Glass (thickness no. 1.5H, Marienfeld, catalogue no. 107172) coated with poly-l-lysine (Sigma, catalogue no. P4832). On the stage, the samples were diluted twofold to obtain a final concentration of 50–100 nM, producing counts around 5,000–6,000. Data were collected using a Refeyn TwoMP mass photometer with AcquireMP v.2024-R2.1. The instrument was focused using droplet dilution. DiscoverMP v.2024-R2.1 was used for Gaussian fitting and generating the distribution of molecular species.
Dynamic light scattering analysis
Dynamic light scattering measurements were performed using Prometheus Panta. RNA samples were folded as above. Two technical replicates were collected using 10-µl capillaries (NanoTemper, catalogue no. PR-C002). Data acquisition was performed for 15 s per capillary at full (100%) laser power and a refractive index of 1.55, using PR.PantaControl software v.1.7.4. The autocorrelation function was calculated, and size distribution data were derived using the default analysis settings in PR.PantaAnalysis v.1.7.4. The size distribution figure was made using GraphPad Prism v.9.0.
Cryo-EM sample preparation
The refolded reaction was mixed with approximately 25 µM total yeast tRNA (Invitrogen, catalogue no. AM7120G) to obtain the datasets resulting in ROOLEfa and ROOLFirm octamers (Extended Data Figs. 2 and 3). Omitting the tRNA resulted in the third dataset with an increased population of ROOLEfa monomers (Extended Data Fig. 4). We hypothesize that the addition of tRNA can shift the equilibrium between the octamer and monomers by affecting the concentrations of cations chelated by RNA or by another mechanism. The mixture was then rapidly chilled on ice and concentrated to approximately 1 μM using an Amicon centrifugal column (MilliporeSigma). A total of 3 μl of the refolded RNA was applied onto glow-discharged (using 15 mA current with negative polarity for 10 s) 200-mesh R1.2/1.3 +2 nm C layer Quantifoil Cu grids. The grids were blotted for 1 s for ROOLFirm and ROOLEfa octamer datasets and 1.5 s for the ROOLEfa monomer dataset at 4 °C and 100% humidity with no blotting offset and rapidly frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific).
Cryo-EM data collection and image processing
Cryo-EM data were acquired using a Titan Krios microscope and Krios G4 (Thermo Fisher Scientific) operating at 300 kV. The Titan Krios microscope at UMass Chan Medical School was equipped with a K3 summit direct detector (Gatan) and a GIF quantum energy filter (Gatan) set to a slit width of 20 eV. The Titan Krios G4 microscope at the Electron Microscopy Center at Fudan University was equipped with a Falcon 4i summit direct detector (Thermo Fisher Scientific). Automated data collection was performed using the EPU v.3.8.1 (ref. 36) and SerialEM v.3.6 (ref. 37) with the beam-image shift method38.
Micrograph series (videos) for ROOLEfa and ROOLFirm octamers were captured in the super-resolution mode at a nominal magnification of ×100,000 resulting in the physical pixel size of 0.83 Å. Videos were collected using the defocus range from −0.8 μm to −1.8 μm. Video stacks were dose fractionated into 40 frames, with a total exposure dose of 51.23 e−/Å2. Videos for the ROOLEfa monomer were captured in super-resolution mode at a nominal magnification of ×96,000 resulting in a physical pixel size of 1.06 Å. Videos were collected using the defocus range from −1.2 μm to −2.2 μm. Video stacks were dose fractionated into 40 frames, with a total exposure dose of 52 e−/Å2.
Cryo-EM datasets were processed using cryoSPARC v.4.6.0 (ref. 39) (Extended Data Figs. 2–4). Initial steps involved patch motion correction and patch contrast transfer function estimation to rectify stage shift, beam-induced motion and imaging distortions. High-quality images were selected for further processing on the basis of ice thickness, defocus ranges and estimated resolutions. Particles were picked using blob picking, followed by two rounds of reference-free 2D classification. Classes containing well-defined particles were re-extracted without binning. Subsequently, a subset of high-quality particles from well-resolved 2D classes underwent ab initio reconstruction and heterogeneous refinement, yielding initial references for both octamers and ROOLEfa monomer. To pick isolated tetrameric particles, 20-Å low pass-filtered tetramer maps obtained from the corresponding octamers were used as a reference. Through homogeneous refinement and reference-based motion correction, followed by non-uniform refinement and local/global refinement, high-resolution maps were obtained for the octamers and ROOLEfa monomer. The ROOLFirm dataset yielded a 3.59-Å resolution map corresponding to the tetramer, whereas the ROOLEfa dataset did not result in a higher resolution reconstruction, probably because the ROOLEfa dataset was smaller than that for ROOLFirm (see octamer particle numbers in Extended Data Table 1). At the final refinement stages, D4 symmetry was applied to ROOLFirm and ROOLEfa octamers, and C4 symmetry to the ROOLFirm tetramer. Final refined maps were sharpened using EMReady v.2.0 beta40. Visualization and evaluation of 3D density maps were performed with UCSF Chimera v.1.16 and ChimeraX v.1.7.1 (ref. 41).
Cryo-EM model building and refinement
We modelled ROOLFirm de novo using Alphafold3 (ref. 29) models of several helical elements clearly resolved in the map, and the rest of the structure was manually built with Coot v.0.9.8.95 (ref. 42) and secondary structure Alphafold3 predictions. The ROOLEfa model was built using secondary structure similarity with ROOLFirm. The monomers were expanded into the corresponding octamer maps. Ions were manually traced into the octamer maps of ROOLEfa and ROOLFirm. Structural models were refined against cryoSPARC maps that were processed in EMready (without supplementing coordinates as input) and sharpened by applying the B factor of −50 Å2 using bfactor.exe as implemented in the Frealign package43. All three models were initially refined using rigid-body refinement in Phenix.real_space_refine44 v.1.19.2. Coordinates were further refined with ISOLDE45 as implemented in ChimeraX41 with added hydrogens and with simulation runtime temperature set to 30. Final individual coordinate minimizations (non-crystallographic symmetry-restrained for both octamers) and atomic displacement parameter refinements were performed in Phenix.real_space_refine v.1.19.2 for ROOLEfa octamer and ROOLEfa monomer and v.1.20.1 for ROOLFirm octamer, yielding average model–map correlation coefficients (CCmask) from 0.78 to 0.90. Final models exhibit good stereochemistry validated by phenix and MolProbity46 online server v.4.5.2 (Extended Data Table 1). Figures were prepared in PyMOL v.3.1.3.1 (ref. 47) and ChimeraX v.1.7.1 (ref. 41). Secondary structure diagrams were drawn with RNArtist (https://github.com/fjossinet/RNArtist) and manually curated with Adobe Illustrator.
Negative-stain electron microscopy sample preparation and data collection
Three drops of 20 μl, 20 μl and 60 μl 1% uranyl acetate stain solution were applied on a parafilm. A 3-μl sample was applied on the glow-discharged (45 s) 300-mesh Cu grids (Quantifoil) coated with a continuous carbon film and incubated for 60 s to enable sample adsorption. The grid was blotted from the side with a piece of filter paper and washed with 3 μl washing buffer. Excessive washing buffer was blotted with filter paper and the grid was stained in the first two drops of uranyl acetate followed by blotting with filter paper. Subsequently, the grid was stained in the third drop of uranyl acetate for 40 s. Excessive uranyl acetate was blotted and the grid was air dried and stored until imaging. All images were acquired with a Talos L120C TEM microscope (FEI Thermo Fisher Scientific) at an accelerating voltage of 120 kV equipped with a Ceta CMOS 4k × 4k pixel camera (FEI Thermo Fisher Scientific) under a magnification of ×96,000 (corresponding to a calibrated sampling of 1.613 Å per physical pixel).
Disease association analysis
Bacterial species and groups encoding ROOL RNAs in the Rfam database48 (https://rfam.org/family/RF03087#tabview=tab4) were searched on the Human Gut Microbiome Atlas website (https://www.microbiomeatlas.org), and their enrichment scores for each listed human disease are plotted in Fig. 1a.
Statistical analysis
All experiments were independently repeated at least once, with no inconsistent results observed. GraphPad Prism v.9.0, OriginLab 2025 and ImageJ v.1.54g were used to perform statistical analyses. Data are presented as mean ± s.d. The details of replicates are described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM maps and associated atomic coordinate models of ROOLEfa octamer, ROOLFirm octamer and ROOLEfa monomer have been deposited in the wwPDB OneDep System under EMD accession codes EMD-48386, EMD-48389 and EMD-48391 and PDB codes 9MM6, 9MME and 9MMG, respectively. The map of ROOLFirm tetramer has been deposited in the EMDB System under EMD accession code EMD-49873. Source data for mass photometry and electron microscopy micrographs associated with Fig. 4e are available at Figshare (https://doi.org/10.6084/m9.figshare.29142887)35. Source data are provided with this paper.
Change history
19 December 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41586-025-10022-0
References
Weinberg, Z., Perreault, J., Meyer, M. M. & Breaker, R. R. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 (2009).
Weinberg, Z. et al. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 45, 10811–10823 (2017).
Harris, K. A. & Breaker, R. R. Large noncoding RNAs in bacteria. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.RWR-0005-2017 (2018).
Pyle, A. M. Group II intron self-splicing. Annu. Rev. Biophys. 45, 183–205 (2016).
Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. The signal recognition particle. Annu. Rev. Biochem. 70, 755–775 (2001).
Stav, S. et al. Genome-wide discovery of structured noncoding RNAs in bacteria. BMC Microbiol. 19, 66 (2019).
Narunsky, A. et al. The discovery of novel noncoding RNAs in 50 bacterial genomes. Nucleic Acids Res. 52, 5152–5165 (2024).
Cousin, F. J. et al. A long and abundant non-coding RNA in Lactobacillus salivarius. Microb. Genom. 3, e000126 (2017).
Bohdan, D. R., Voronina, V. V., Bujnicki, J. M. & Baulin, E. F. A comprehensive survey of long-range tertiary interactions and motifs in non-coding RNA structures. Nucleic Acids Res. 51, 8367–8382 (2023).
Cate, J. H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Doherty, E. A., Batey, R. T., Masquida, B. & Doudna, J. A. A universal mode of helix packing in RNA. Nat. Struct. Mol. Biol. 8, 339–343 (2001).
Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B. & Steitz, T. A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA 98, 4899–4903 (2001).
Battle, D. J. & Doudna, J. A. Specificity of RNA–RNA helix recognition. Proc. Natl Acad. Sci. USA 99, 11676–11681 (2002).
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Demeshkina, N., Jenner, L., Westhof, E., Yusupov, M. & Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012).
Loveland, A. B., Demo, G., Grigorieff, N. & Korostelev, A. A. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117 (2017).
Teran, D., Zhang, Y. & Korostelev, A. A. Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon. Front. Microbiol. https://doi.org/10.3389/fmicb.2024.1369760 (2024).
Autour, A. et al. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 9, 656 (2018).
Trachman, R. J. et al. Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat. Chem. Biol. 15, 472–479 (2019).
Albers, S. et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 618, 842–848 (2023).
Record, M. T. Jr, Zhang, W. & Anderson, C. F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 51, 281–353 (1998).
Shiman, R. & Draper, D. E. Stabilization of RNA tertiary structure by monovalent cations. J. Mol. Biol. 302, 79–91 (2000).
Szatmári, D. et al. Intracellular ion concentrations and cation-dependent remodelling of bacterial MreB assemblies. Sci. Rep. 10, 12002 (2020).
Greening, C. & Lithgow, T. Formation and function of bacterial organelles. Nat. Rev. Microbiol. 18, 677–689 (2020).
Chen, A. G. Functional Investigation of Ribozymes and Ribozyme Candidates in Viruses, Bacteria and Eukaryotes. PhD thesis, Yale Univ. (2015).
Kretsch, R. C. et al. Naturally ornate RNA-only complexes revealed by cryo-EM. Nature https://doi.org/10.1038/s41586-025-09073-0 (2025).
Patel, S. D. et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124, 1255–1268 (2006).
Häge, F. R. et al. Strand-swapped SH3 domain dimer with superoxide dismutase activity. ACS Cent. Sci. 11, 157–166 (2025).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021).
Jasinski, D., Haque, F., Binzel, D. W. & Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 11, 1142–1164 (2017).
Shu, Y. et al. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 19, 767–777 (2013).
Høiberg, H. C., Sparvath, S. M., Andersen, V. L., Kjems, J. & Andersen, E. S. An RNA origami octahedron with intrinsic siRNAs for potent gene knockdown. Biotechnol. J. 14, e1700634 (2019).
Afonin, K. A., Dobrovolskaia, M. A., Ke, W., Grodzinski, P. & Bathe, M. Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Deliv. Rev. 181, 114081 (2022).
Ling, X., Golovenko, D., Gan, J., Ma, J., Korostelev, A. A. & Fang, W. Cryo-EM structure of a natural RNA nanocage. Dataset. Figshare https://doi.org/10.6084/m9.figshare.29142887 (2025).
Thompson, R. F., Iadanza, M. G., Hesketh, E. L., Rawson, S. & Ranson, N. A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Wu, C., Huang, X., Cheng, J., Zhu, D. & Zhang, X. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol. 208, 107396 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
He, J., Li, T. & Huang, S.-Y. Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Grigorieff, N. Frealign: an exploratory tool for single-particle cryo-EM. Methods Enzymol. 579, 191–226 (2016).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
The PyMOL Molecular Graphics System v.3.1.3.1 (Schrödinger, LLC, 2010).
Ontiveros-Palacios, N. et al. Rfam 15: RNA families database in 2025. Nucleic Acids Res. https://doi.org/10.1093/nar/gkae1023 (2024).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Acknowledgements
Cryo-EM data were collected at the Electron Microscopy Center at Fudan University and the Cryo-EM Core Facility at UMass Chan Medical School. The data were processed at the High Performance Computing Center of the Electron Microscopy Center at Fudan University and the HPC cluster at UMass Chan Medical School. We thank K. Song and C. Ouch at the Cryo-EM Core Facility at UMass Chan Medical School for assistance with collecting 300 kV cryo-EM data; K. Lee at the EM Facility at UMass Chan Medical School for collecting negative-stain EM data; L. Robbins and C. Hull (UMass Chan Medical School) for assistance with HPC usage; D. Zhu (Howard Hughes Medical Institute) and J. Zhang (Harvard University) for assistance with data processing; S. Klinge (Rockefeller University) and T. Li (Huazhong University of Science and Technology) for assistance with modelling; B. Kelch, R. Ahsan, L. Li and Y. A. Nguyen (UMass Chan Medical School) for assistance with ROOL characterization in solution; K. Lowenhaupt at the Biophysical Instrumentation Facility (Massachusetts Institute of Technology) for assistance with mass photometry and dynamic light scattering experiments; Z. Weinberg (Martin Luther University Halle-Wittenberg) for helpful discussions; A. LueCheeLip (UMass Chan Medical School) for technical assistance; and D. Conte Jr (UMass Chan Medical School) for editing the manuscript. This work was supported by National Natural Science Foundation of China (NSFC32471347 to J.M.), Program of Shanghai Academic/Technology Research Leader (to J. M.), National Institutes of Health, National Institute of General Medical Sciences (R35GM127094 to A.A.K. and R35GM150953 to W.F.) and UMass Chan Medical School Startup fund to W.F.
Author information
Authors and Affiliations
Contributions
X.L. conceived the project. J.M., A.A.K. and W.F. supervised the project and participated in experimental design and analysis. X.L. performed experiments and analysed data, prepared cryo-EM samples and collected and processed cryo-EM data. X.L., D.G., J.G., J.M. and A.A.K. built, refined and validated atomic coordinate models. X.L., D.G., A.A.K. and W.F. prepared the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The University of Massachusetts has filed a provisional patent application based on the innovation disclosed herein.
Peer review
Peer review information
Nature thanks Anna Pyle and Jane Richardson for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Biochemical analyses of ROOL RNAs.
a, Denaturing PAGE analyses demonstrate that the RNAs are intact full-length molecules. RNAs were analysed at least once. b, Mass photometry analysis of ROOLEfa and ROOLFirm at 50–100 nM in 240 mM K+ and 20 mM Mg2+ shows that monomers and octamers are the two most abundant species. The RNA size of each peak was calibrated using the Millennium RNA Marker and labelled. c, Size exclusion chromatography (SEC) analyses of ROOLEfa and ROOLFirm coupled with denaturing PAGE (d), mass photometry (e), and negative-stain EM (f) showed three major states (aggregates, octamer, and monomer) in the cryo-EM samples. Peak 1 (void volume) contained aggregated ROOL of different oligomerization states; peaks 2 and 3 correspond to predominantly octamers and monomers, respectively. Experiments in c and e were repeated at least once with similar results; experiments in d and f were performed once; three EM micrographs were analysed in f. g, Dynamic light scattering shows a diameter range of 10–30 nm for ROOL RNAs, consistent with its oligomerization states. A representative result from two independent technical replicates is shown. For original RNA gel images and negative-stain micrographs, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM data processing workflow for ROOLEfa.
a, Maximum-likelihood classification of cryo-EM data using cryoSPARC. b, Example of a micrograph with ROOLEfa particles. 6,966 micrographs were obtained and analysed. c, Fourier shell correlation curve as a function of resolution for the final map shown in panels a and e. d, Euler angle distribution of particles contributing to the final reconstruction. e, Cryo-EM maps coloured according to local resolution.
Extended Data Fig. 3 Cryo-EM data processing workflow for ROOLFirm.
a, Maximum-likelihood classification of cryo-EM data using cryoSPARC. b, Example of a micrograph with ROOLFirm particles. 6,390 micrographs were obtained and analysed. c, Fourier shell correlation curves as a function of resolution for the final octamer and tetramer maps shown in panels e and f. d, Euler angle distribution of particles contributing to the final reconstructions. e, f, Cryo-EM maps of the octamer (e) and tetramer (f), coloured according to local resolution.
Extended Data Fig. 4 Cryo-EM data processing workflow for the individual monomer of ROOLEfa.
a, Maximum-likelihood classification of cryo-EM data using cryoSPARC. b, Example of a micrograph with ROOLEfa particles. 5,253 micrographs were obtained and analysed. c, Cryo-EM maps coloured according to local resolution. d, Fourier shell correlation curves as a function of resolution for the final maps shown in panels a and c. e, Euler angle distribution of particles contributing to the final reconstruction.
Extended Data Fig. 5 Tertiary interactions stabilize each ROOLFirm monomer in the nanocage.
a, Molecular model of a ROOLFirm monomer within the octameric structure. Close-up views of examples of tertiary interactions stabilizing the monomer are shown in the corresponding coloured boxes in panels b–g with map rendered as transparent surface (σ = 4.16). Panels b and f show different views for the orange box in panel a. h, Secondary structure annotation of ROOLEfa within the octamer, with tertiary interactions labelled by coloured boxes that match panels a–g. Nucleotides in panels b–h are coloured according to helix colours in a.
Extended Data Fig. 6 Projected secondary structure with tertiary and quaternary interactions of ROOLEfa shown based on the cryo-EM structure.
H1–H16 of monomers 1–4, H7, and H13, H15, H16 of monomers 5–8 are shown to indicate tertiary and quaternary interactions in ROOLEfa. In ROOLFirm, G-A stacking replaces G-C Hoogsteen base pairing.
Extended Data Fig. 7 Quaternary interactions in ROOLFirm and inter-tetramer interactions in ROOLFirm and ROOLEfa.
a, Front view of the ROOLFirm dimer stabilized by four key interactions, the close-up views of which are shown in panels c–f with map rendered as transparent surface (σ = 4.16). b, Side view of the inter-tetramer interface of ROOLFirm, the close-up views of which are shown in panels g and h with map rendered as transparent surface (σ = 4.16). i, Side view of the inter-tetramer interface of ROOLEfa (viewed similarly to ROOLFirm in b), the close-up views of which are shown in panels j and k with map rendered as transparent surface (σ = 2.88).
Extended Data Fig. 8 Mutational analysis of H12 supports the strand-swapping mechanism.
a, Predicted secondary structures (by UNAFold49) of the WT or mutated H12 sequences of ROOLEfa. Nucleotides that participate in new kissing-loop interactions with H2 and H11 in the octamer are circled in red and green boxes, respectively. The purple shade indicates the disorder region in the octamer structure. In mut1, a G is inserted after U356 to pair with C423; in mut2, AAU (417–419) is changed to a C to pair with G360; in mut3, the stem-loop structure in the disordered region was replaced by a more stable hairpin (GC-rich with a GAAA tetra loop); mut4 combines the mutations in mut1 and mut2; mut5 combines mut4 and an additional removal of a bulge shown in dark blue on the WT structure diagram. b, c, Negative-stain EM analysis of the WT and mutant ROOLEfa. Representative micrographs are shown in b and quantifications from three images for each sample are shown in c. Data are presented as mean ± SD. For original negative-stain micrographs, see Supplementary Fig. 1. d, SEC analyses of WT and mutant ROOLEfa constructs. Two independent experiments were performed yielding similar results, and a representative chromatogram is shown for each construct. e, Mass photometry analysis of ROOLEfa H12 mutants. Two technical replicates were performed, yielding similar results.
Extended Data Fig. 9 Analysis of ROOLEfa oligomerization at varying salt concentrations.
a, Negative-stain EM analysis of ROOLEfa at different K+ and Mg2+ concentrations. b, Quantification of ROOLEfa nanocage formation using three different full images from the negative-stain EM analysis of each condition shown in a. Data are presented as mean ± SD. For original negative-stain micrographs, see Supplementary Fig. 1. c, SEC analyses of ROOLEfa show that different K+ and Mg2+ concentrations shift the distributions among three major peaks/states (aggregates, octamer, and monomer). Two independent experiments were performed yielding similar results, and a representative chromatogram is shown for each condition. d, Mass photometry analysis of ROOLEfa at different K+ and Mg2+ concentrations. Two technical replicates were performed, yielding similar results.
Supplementary information
Supplementary Fig. 1
Original gel images and negative-stain micrographs. These include original gel images associated with Extended Data Fig. 1a,d; original negative-stain micrographs associated with Fig. 4c,e and Extended Data Figs. 1f, 8b,c and 9a,b.
Supplementary Table 1
Nucleotide sequences used in the study.
Supplementary Video 1
Structure of ROOLEfa octamer. The overall architecture and important interactions are featured.
Supplementary Video 2
Structure of ROOLEfa monomer and the strand-swapping mechanism. The overall structure of ROOLEfa monomer, its alignment with ROOLEfa monomer within the octamer and the strand-swapping mechanism are featured.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ling, X., Golovenko, D., Gan, J. et al. Cryo-EM structure of a natural RNA nanocage. Nature 644, 1107–1115 (2025). https://doi.org/10.1038/s41586-025-09262-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09262-x
This article is cited by
-
Integrated experimental and AI innovations for RNA structure determination
Nature Biotechnology (2026)
-
Decoding the structure and translational promise of ROOL RNA nanocages
Molecular Biomedicine (2025)