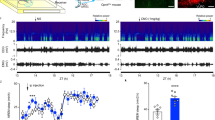
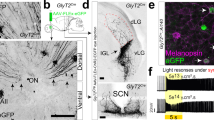
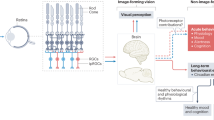
Abstract
The suprachiasmatic nucleus (SCN), the central circadian pacemaker, receives photic input exclusively from intrinsically photosensitive retinal ganglion cells (ipRGCs)1,2. However, light mainly shifts the SCN clock during night-time3,4,5. Here we induced phase shifts in the SCN clock during the daytime in mice by activating ipRGCs using chemogenetics or violet light. Our data reveal that the inability to induce daytime shifts with light in most animals is not only attributed to the SCN, as has been proposed for decades, but also requires the limitation of ipRGC firing via depolarization block. Chemogenetic activation of ipRGCs induces large shifts during both night-time and daytime, but daytime shifts require brain circuits and neuropeptide transmitters that are dispensable for night-time shifts. Thus, propensity of ipRGCs for depolarization block not only prevents daytime shifts in mice, but also limits the magnitude of night-time shifts, suggesting that ipRGC inputs to SCN act as an integrated pacemaker across the circadian cycle.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The principal data supporting the findings of this Article are available within the figures and the Supplementary Information; additional data that support the findings of this study are available from the corresponding authors on request.
Code availability
Custom code used to analyse electrophysiological data is available at http://www.github.com/SchwartzNU/SymphonyAnalysis.
References
Hattar, S., Liao, H. W., Takao, M., Berson, D. M. & Yau, K. W. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070 (2002).
Jones, J. R., Simon, T., Lones, L. & Herzog, E. D. SCN VIP neurons are essential for normal light-mediated resetting of the circadian system. J. Neurosci. 38, 7986–7995 (2018).
de Vries, M. J., Treep, J. A., de Pauw, E. S. & Meijer, J. H. The effects of electrical stimulation of the optic nerves and anterior optic chiasm on the circadian activity rhythm of the Syrian hamster: involvement of excitatory amino acids. Brain Res. 642, 206–212 (1994).
Ding, J. M. et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713 (1994).
Mintz, E. M., Marvel, C. L., Gillespie, C. F., Price, K. M. & Albers, H. E. Activation of NMDA Receptors in the suprachiasmatic nucleus produces light-like phase shifts of the circadian clock in vivo. J. Neurosci. 19, 5124–5130 (1999).
Contreras, E., Nobleman, A. P., Robinson, P. R. & Schmidt, T. M. Melanopsin phototransduction: beyond canonical cascades. J. Exp. Biol. 224, jeb226522 (2021).
Milner, E. S. & Do, M. T. H. A population representation of absolute light intensity in the mammalian retina. Cell 171, 865–876.e816 (2017).
Do, M. T. H. Melanopsin and the intrinsically photosensitive retinal ganglion cells: biophysics to behavior. Neuron 104, 205–226 (2019).
Chen, S.-K., Badea, T. & Hattar, S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95 (2011).
Vitaterna, M. H., Takahashi, J. S. & Turek, F. W. Overview of circadian rhythms. Alcohol Res. Health 25, 85 (2001).
Johnson, C. H. Circadian Clocks from Cell to Human (eds T. Hiroshige, T. & Honma, K.) 209–249 (Hokkaido Univ. Press, 1992).
COURSEY, P. J. D. Daily light sensitivity rhythm in a rodent. Science 131, 33–35 (1960).
Kornhauser, J. M., Ginty, D. D., Greenberg, M. E., Mayo, K. E. & Takahashi, J. S. Light entrainment and activation of signal transduction pathways in the SCN. Prog. Brain Res. 111, 133–146 (1996).
Shirakawa, T. & Moore, R. Y. Glutamate shifts the phase of the circadian neuronal firing rhythm in the rat suprachiasmatic nucleus in vitro. Neurosci. Lett. 178, 47–50 (1994).
Asai, M. et al. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr. Biol. 11, 1524–1527 (2001).
Jones, J. R., Tackenberg, M. C. & McMahon, D. G. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat. Neurosci. 18, 373–375 (2015).
Kim, S. & McMahon, D. G. Light sets the brain’s daily clock by regional quickening and slowing of the molecular clockworks at dawn and dusk. eLife 10, e70137 (2021).
Rusak, B. & Groos, G. Suprachiasmatic stimulation phase shifts rodent circadian rhythms. Science 215, 1407–1409 (1982).
Shibata, S. & Moore, R. Y. Neuropeptide Y and optic chiasm stimulation of affect suprachiasmatic nucleus circadian function in vitro. Brain Res. 615, 95–100 (1993).
Shibata, S., Watanabe, A., Hamada, T., Ono, M. & Watanabe, S. N-methyl-D-aspartate induces phase shifts in circadian rhythm of neuronal activity of rat SCN in vitro. Am. J. Physiol. 267, R360–R364 (1994).
Sladek, M. & Sumova, A. Modulation of NMDA-mediated clock resetting in the suprachiasmatic nuclei of mPer2Luc mouse by endocannabinoids. Front. Physiol. 10, 361 (2019).
Gillette, M. U. & Mitchell, J. W. Signaling in the suprachiasmatic nucleus: selectively responsive and integrative. Cell Tissue Res. 309, 99–107 (2002).
Yan, L., Foley, N. C., Bobula, J. M., Kriegsfeld, L. J. & Silver, R. Two antiphase oscillations occur in each suprachiasmatic nucleus of behaviorally split hamsters. J. Neurosci. 25, 9017–9026 (2005).
Liu, A. et al. Encoding of environmental illumination by primate melanopsin neurons. Science 379, 376–381 (2023).
Atasoy, D. & Sternson, S. M. Chemogenetic tools for causal cellular and neuronal biology. Physiol. Rev. 98, 391–418 (2018).
van Diepen, H. C. et al. Distinct contribution of cone photoreceptor subtypes to the mammalian biological clock. Proc. Natl Acad. Sci. USA 118, e2024500118 (2021).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Ecker, J. L. et al. Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67, 49–60 (2010).
Beier, C., Zhang, Z., Yurgel, M. & Hattar, S. Projections of ipRGCs and conventional RGCs to retinorecipient brain nuclei. J. Comp. Neurol. 529, 1863–1875 (2021).
Duy, P. Q. et al. Light has diverse spatiotemporal molecular changes in the mouse suprachiasmatic nucleus. J. Biol. Rhythms 35, 576–587 (2020).
Engelund, A., Fahrenkrug, J., Harrison, A. & Hannibal, J. Vesicular glutamate transporter 2 (VGLUT2) is co-stored with PACAP in projections from the rat melanopsin-containing retinal ganglion cells. Cell Tissue Res. 340, 243–255 (2010).
Keenan, W. T. et al. A visual circuit uses complementary mechanisms to support transient and sustained pupil constriction. eLife 5, e15392 (2016).
Harrington, M. E. The ventral lateral geniculate nucleus and the intergeniculate leaflet: interrelated structures in the visual and circadian systems. Neurosci. Biobehav. Rev. 21, 705–727 (1997).
Card, J. P. & Moore, R. Y. Organization of lateral geniculate-hypothalamic connections in the rat. J. Comp. Neurol. 284, 135–147 (1989).
Monavarfeshani, A., Sabbagh, U. & Fox, M. A. Not a one-trick pony: Diverse connectivity and functions of the rodent lateral geniculate complex. Vis. Neurosci. 34, E012 (2017).
Sabbah, S. et al. Intrinsically photosensitive retinal ganglion cells evade temporal filtering to encode environmental light intensity. Preprint at bioRxiv https://doi.org/10.1101/2022.04.09.487733 (2022).
Haverkamp, S. et al. The primordial, blue-cone color system of the mouse retina. J. Neurosci. 25, 5438–5445 (2005).
Behrens, C., Schubert, T., Haverkamp, S., Euler, T. & Berens, P. Connectivity map of bipolar cells and photoreceptors in the mouse retina. eLife 5, e20041 (2016).
Nadal-Nicolás, F. M. et al. True S-cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field. eLife 9, e56840 (2020).
Iyer, R., Wang, T. A. & Gillette, M. U. Circadian gating of neuronal functionality: a basis for iterative metaplasticity. Front. Syst. Neurosci. 8, 164 (2014).
Yan, L., Smale, L. & Nunez, A. A. Circadian and photic modulation of daily rhythms in diurnal mammals. Eur. J. Neurosci. 51, 551–566 (2020).
Raper, J. et al. Metabolism and distribution of clozapine-N-oxide: implications for nonhuman primate chemogenetics. ACS Chem. Neurosci. 8, 1570–1576 (2017).
Lindberg, P. T. et al. Pituitary adenylate cyclase-activating peptide (PACAP)-glutamate co-transmission drives circadian phase-advancing responses to intrinsically photosensitive retinal ganglion cell projections by suprachiasmatic nucleus. Front. Neurosci. 13, 484713 (2019).
Blume, C., Garbazza, C. & Spitschan, M. Effects of light on human circadian rhythms, sleep and mood. Somnologie 23, 147 (2019).
Jewett, M. E. et al. Human circadian pacemaker is sensitive to light throughout subjective day without evidence of transients. Am. J. Physiol. 273, R1800–R1809 (1997).
Revell, V. L., Molina, T. A. & Eastman, C. I. Human phase response curve to intermittent blue light using a commercially available device. J. Physiol. 590, 4859–4868 (2012).
Mure, L. S. Intrinsically photosensitive retinal ganglion cells of the human retina. Front. Neurol. 12, 636330 (2021).
Ross, R. A. et al. PACAP neurons in the ventral premammillary nucleus regulate reproductive function in the female mouse. eLife 7, e35960 (2018).
van Norren, D. & Gorgels, T. G. The action spectrum of photochemical damage to the retina: a review of monochromatic threshold data. Photochem. Photobiol. 87, 747–753 (2011).
Henriksson, J. T., Bergmanson, J. P. & Walsh, J. E. Ultraviolet radiation transmittance of the mouse eye and its individual media components. Exp. Eye Res. 90, 382–387 (2010).
Jeon, C.-J., Strettoi, E. & Masland, R. H. The major cell populations of the mouse retina. J. Neurosci. 18, 8936–8946 (1998).
Govardovskii, V. I., Calvert, P. D. & Arshavsky, V. Y. Photoreceptor light adaptation: untangling desensitization and sensitization. J. Gen. Physiol. 116, 791–794 (2000).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Nath, A., Grimes, W. N. & Diamond, J. S. Layers of inhibitory networks shape receptive field properties of AII amacrine cells. Cell Rep. 42, 113390 (2023).
Franke, K. et al. An arbitrary-spectrum spatial visual stimulator for vision research. eLife 8, e48779 (2019).
Acknowledgements
We thank the members of the SLCR at NIMH and the Johns Hopkins Biology Mouse Tri-Lab for helpful discussions. Funding sources: National Institute of Mental Health Grant MH002964 (S.H.), K99 Grant EY033414-01 (C.B.), National Institute of Mental Health Grant MH002950 (M.P.), NIMH IRP Rodent Behavioral Core (MH002952), NIH grant EY027202 (H.Z. and R.L.B), NIH grant EY29985 and NINDS Intramural Research Program NS003145 (J.S.D.).
Author information
Authors and Affiliations
Contributions
R.K., C.B. and S.H. conceptualized the study. R.K. and S.Y. collected immunohistochemistry data. R.K., C.B., H.W. and C.G. collected behavioural data. R.K., C.B., C.G., M.P. and S.H. designed behavioural experiments. A.N., W.N.G., G.P.O., R.L.B. and J.S.D. designed, performed and analysed the electrophysiology experiments. M.B. and B.S. designed, performed and analysed MEA recording experiments. R.K., C.B., M.T., S.Y. and H.W. generated and maintained mouse crosses used in the study. R.K., C.B., A.N., W.N.G. and H.Z. analysed the data. R.K., C.B., W.N.G., H.Z. and S.H. wrote the manuscript. R.K., C.B., A.N., W.N.G., G.P.O., J.B., W.L., R.L.B., J.D., H.Z. and S.H. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Chemogenetic activation of ipRGCs induces acute H3 phosphorylation and cFos expression in the SCN at CT6, daytime phase shift in the circadian clock and depolarization block in ipRGC. Related to Figs. 1, 2, 3.
a. Immunohistochemistry staining shows that long duration (1, 3, or 6 h) light pulse ending at CT6 does not induce H3 phosphorylation in the SCN. Scale bars 100 µm. b. Quantification of H3 phosphorylation in the SCN following treatments in a. Long light pulses (1-, 3-, and 6-h, n = 3 per condition) do not induce significant changes in H3 phosphorylation (p ≥ 0.9804, one way ANOVA). Plotted data shows mice as individual points with mean ± SEM. **** p < 0.0001, and ns is not significant p > 0.05. Data from Fig.1 (CNO + LP; Saline + LP) are replotted for comparison. c. Immunohistochemistry staining shows that chemogenetic activation of ipRGCs without light pulse induced H3 phosphorylation (green) in the SCN at CT6. The SCN (dashed outline) was visualized with DAPI (blue). ipRGC projections are visible with mCherry (magenta) within the SCN. Scale bars 100 µm. d. CNO injection and light pulse (LP) at CT6 in Opn4Cre mice without hM3D(Gq) did not induce H3 phosphorylation (green). e. Quantification of H3 phosphorylation cell density in the SCN shown in c and d. Data from Fig.1 (CNO+LP) is replotted for comparison. CT6 CNO injections in Gq-Opn4Cre mice induce H3 phosphorylation with (n=5) or without light pulse (n=5). Plotted data shows mice as individual points with mean ± SEM. p = 0.4001, one way ANOVA. H3 phosphorylation was significantly smaller in mice without hM3D(Gq) after CNO injection and light pulse at CT6 (p ≤ 0.001, one way ANOVA). f. Immunohistochemistry staining shows the cFos expression (green) in the SCN at CT6 in response to saline injection and light pulse. The SCN (dashed outline) was visualized with DAPI (blue). ipRGC projections are visible with mCherry (magenta) within the SCN. Scale bars 100 µm. g. Same as in f, but in response to chemogenetic activation. Scale bars 100 µm. h. Quantification of cFos expression cell density in the SCN shown in f and g. cFos expression cell density was significantly greater after a CNO injection (n=5) compared to saline injection (n=4) in Gq-Opn4Cre mice. Plotted data shows mice as individual points with mean ± SEM. p < 0.0001, unpaired two-tailed t-test). i. The double-plotted actograms of all wild type mice (n = 4). Wheel running activity is indicated by dark bars. On day 8 the mice received a 1-h light pulse (LP) from CT5- CT6. j. The double-plotted actograms of all wild type mice (n = 5). On day 8 the mice received a 3-h light pulse (LP) from CT3- CT6. k. The double-plotted actograms of all wild type mice (n = 5). On day 8 the mice received a 6-h light pulse (LP) from CT0- CT6. l. The double-plotted actograms of all male Gq-Opn4Cre/Cre mice (n = 4). Wheel running activity is indicated by dark bars. On day 8 the mice received a saline injection at CT4 and a 15-min light pulse (LP) at CT6. m. The double-plotted actograms of all male Gq-Opn4Cre/Cre mice (n = 4). On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. n. The double-plotted actograms of all female Gq-Opn4Cre/Cre mice (n = 4). On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. o. The double-plotted actograms of all Gq-Opn4Cre/+ mice (n = 5). On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. p. Quantification of CNO-induced phase shifts observed in m-o. Phase shifts in male Gq-Opn4Cre/Cre mice (n=5), female Gq-Opn4Cre/Cre mice (n=3), and Gq-Opn4Cre/+ mice (n=5) are not significantly different (p ≥ 0.2225, one way ANOVA). Plotted data shows mice as individual points with mean ± SEM. q. Whole cell current clamp recording from an M1 ipRGC in the dark under control conditions, and in the presence of 5 and 50 µM CNO. The horizontal dashed line indicates the initial resting membrane potential. On the right side, quantification of average spike rates (n = 4 cells) in response to control and two CNO steps. L-AP4 (10 µM) was present in the bath throughout these whole cell recordings, thus isolating contributions from melanopsin.
Extended Data Fig. 2 Actograms for Gq-Opn4Cre;Adcyap1fl mice injected with CNO during the subjective day (a) or subjective night (b).
Related to Figs. 2 and 3. a. The double-plotted actograms of all Gq-Opn4Cre;Adcyap1fl mice (n = 7) during daytime activation of ipRGCs. On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. b. The double-plotted actograms of all Gq-Opn4Cre;Adcyap1fl mice (n = 5) during nighttime activation of ipRGCs. On day 8 the mice received a CNO injection at CT12 and a 15-min LP at CT14. c. Actograms for Gq-Opn4Cre mice injected with CNO during the subjective night. The double-plotted actograms of all Gq-Opn4Cre mice (n = 13) during nighttime activation of ipRGCs. On day 8 the mice received a CNO injection at CT12 and a 15-min LP at CT14. Quantifications of observed phase shifts in a-c are shown in Figs. 2 and 3.
Extended Data Fig. 3 Different circuits are required for daytime and nighttime phase shift.
Related to Fig. 3. a-c. SCN-projecting ipRGCs are not sufficient to induce daytime phase delays. a. Schematic representations of ipRGC projections in Gq-Opn4Cre (top panel) and Gq-Opn4Cre;Brn3bDTA (bottom panel) mice. In Gq-Opn4Cre;Brn3bDTA mice, where the extra-SCN-projecting Brn3b-positive ipRGCs are ablated, the surviving M1Brn3b− ipRGCs predominantly project to the SCN with minor projections to several other brain regions, including the intergeniculate leaflet (IGL), ventral lateral geniculate nucleus (vLGN), and the olivary pretectal nucleus (OPN). b. Immunohistochemistry staining shows that chemogenetic activation of the surviving M1Brn3b− ipRGCs in Gq-Opn4Cre;Brn3bDTA mice (bottom panels) does not induce H3 phosphorylation (green) in the SCN at CT6. The SCN (dashed outline) was visualized with DAPI (blue). ipRGC projections are visible with mCherry (magenta) within the SCN. Scale bars 100 µm. c. Quantification of H3 phosphorylation cell density in the SCN shown in b. H3 phosphorylation cell density was significantly smaller in Gq-Opn4Cre;Brn3bDTA mice (n=4) compared to Gq-Opn4Cre mice (n=5) (p < 0.0001, unpaired two-tailed t-test). Plotted data shows mice as individual points with mean ± SEM. Gq-Opn4Cre data (b and c) are re-shown from Fig. 1 for comparison. d. In Gq-Opn4Cre mice (upper panel) and Gq-Opn4Cre;Brn3bDTA mice (lower panel) ipRGC projections are visible with mCherry (magenta) within the suprachiasmatic nucleus (SCN), the olivary pretectal nucleus (OPN), the lateral geniculate nucleus (LGN), and the superior colliculus (SC). In Gq-Opn4Cre;Brn3bDTA mice ipRGC projections are visible with mCherry within the SCN. Minimal projections are visible in the OPN, LGN, and SC. The remaining projections to the LGN are restricted to the intergeniculate leaflet and the ventral lateral geniculate nucleus. All panels are from one animal. n = 4 mice. Scale bars 100 µm. e-f. Actograms for Gq-Opn4Cre;Brn3bDTA mice injected with CNO during the subjective day and subjective night. e. The double-plotted actograms of all Gq-Opn4Cre;Brn3bDTA mice (n = 9) during daytime activation of ipRGCs. On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. f. The double-plotted actograms of all Gq-Opn4Cre;Brn3bDTA mice (n = 9) during nighttime activation of ipRGCs. On day 8 the mice received a CNO injection at CT12 and a 15-min LP at CT14. Quantifications of observed phase shifts in e-f are shown in Fig. 3. g-m. Silencing of LGN neuron g. Schematic representations of ipRGC projections in Gq-Opn4Cre (top panel) and the LGN-silenced Gq-Opn4Cre mice (bottom panel). h. Immunohistochemistry staining shows the cFos expression (green) in the LGN following chemogenetic activation of ipRGCs in Gq-Opn4Cre (top panel) and the LGN-silenced Gq-Opn4Cre mice (bottom panel). ipRGC projections are visible with mCherry (magenta) within the LGN. Scale bars 100 µm. i. Magnified view of marked LGN in h, cFos expression is visible in green and ipRGC projections are visible with mCherry (magenta) in Gq-Opn4Cre (top panel) and the LGN-silenced Gq-Opn4Cre mice (bottom panel). Scale bars 100 µm. j. Quantification of cFos expression cell density in the LGN shown in i. cFos expression cell density was significantly reduced in the LGN-silenced Gq-Opn4Cre mice (n=3) compared to Gq-Opn4Cre mice (n=3) after a CNO. Plotted data shows mice as individual points with mean ± SEM. p = 0.0020, unpaired two-tailed t-test. k. Schematic representations of silencing of LGN in Opn4Cre mice. l. Immunohistochemistry staining shows the inhibitory DREADD expression that are visible with mCherry (magenta) within the LGN in LGN-silenced Opn4Cre mice. LGN was visualized with DAPI (blue). Scale bars 1000 µm. Magnified view is shown in right. n = 4 mice. Scale bars 100 µm. m. Whole mount retina (Scale bars 1000 µm) immunohistochemistry staining of LGN-silenced Opn4Cre mice. Melanopsin (Opn4) are visible in green. There was no mCherry (magenta) expression was observed in retina. n = 3 mice. Magnified view, Scale bars 100 µm. n-o. Actograms for LGN-silenced Gq-Opn4Cre mice injected with CNO during the subjective day and subjective night. n. The double-plotted actograms of all LGN-silenced Gq-Opn4Cre mice (n = 11) during daytime activation of ipRGCs. On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. o. The double-plotted actograms of all LGN-silenced Gq-Opn4Cre mice (n = 11) during nighttime activation of ipRGCs. On day 8 the mice received a CNO injection at CT12 and a 15-min LP at CT14. Quantifications of observed phase shifts in n-o are shown in Fig. 3. p-s. Chemogenetic activation of only Brn3b-positive ipRGCs. p. The double-plotted actograms of all Gq-Brn3b Cre;Opn4flpo mice (n = 4) during daytime activation of Brn3b-positive ipRGCs. On day 8 the mice received a CNO injection at CT4 and a 15-min LP at CT6. q. Quantifications of observed phase shifts in p and comparison of daytime phase shift by chemogenetic activation of all ipRGCs (Gq-Opn4Cre mice, n = 13) vs only brn3b-positive ipRGCs (Gq-Brn3b Cre;Opn4flpo mice n = 4). Plotted data shows mice as individual points with mean ± SEM. p < 0.0001, unpaired two-tailed t-test. r. Pupillary response after saline and CNO injection in Gq-Brn3b Cre;Opn4flpo mice (n = 3). s. Quantifications of observed PLR in r (n = 3 per condition). Plotted data shows mice as individual points with mean ± SEM. p = 0.0303, paired two-tailed t-test.
Extended Data Fig. 4 Actograms for wild type mice exposed to 15-min of blue/ violet light and example recording showing the responses to sequential staircase protocols.
Related to Fig. 4. a-b. Actograms for wild type mice exposed to 15-min of blue or violet light. a. The double-plotted actograms of 8 wild type mice. On day 8 the mice received a 15-min blue light pulse (LP) at CT6. b. The double-plotted actograms of 8 wild type mice. On day 8 the mice received a 15-min violet light pulse (LP) at CT6. c-h. Representative single-cell recording illustrating the full protocol associated with Fig. 4e–k, including recovery from depolarization block. c. Violet light stimulation protocol including recovery period. d. Continuous raw spike activity in response to violet light stimulation. e. Spike rate histogram (5-s bins) corresponding to the trace in panel d. f. Blue light stimulation protocol including recovery period, which immediately followed the protocol shown in c g. Continuous raw spike activity in response to blue light stimulation. h. Spike rate histogram (5-s bins) corresponding to the trace in panel g. i-k Key statistics in Fig. 4i–k remain consistent regardless of the order of stimulus delivery. p-values are from unpaired t-tests. i. The peak firing rate ratios of violet:blue during the first 5 s of each luminance increment, grouped by LED presentation order. Plotted data show individual cells as points with mean ± SEM. n = 9 cells for violet-first and n = 11 cells for blue-first conditions. p = 0.84, unpaired two-tailed t-test. j. The violet:blue Iblock ratios grouped by LED presentation order. Plotted data show individual cells as points with mean ± SEM. n = 7 cells for violet-first and n = 9 cells for blue-first conditions. p = 0.67, unpaired two-tailed t-test. k. The tuning range ratios of violet:blue, grouped by LED presentation order. Plotted data show individual cells as points with mean ± SEM. n = 7 cells for violet-first and n = 9 cells for blue-first conditions. p = 0.77, unpaired two-tailed t-test. l. The double-plotted actograms of 11 wild-type mice. On day 8 the mice received a 15-min blue light pulse (LP) at CT6. Upper panel: λmax =470 nm; 2.44 × 109 − 1.1 × 1010 photons/cm2/s (n=5 mice); Bottom panel: 1.2 × 10¹¹ − 5.1 × 10¹² photons/cm²/s (n=6 mice). The daily activity onsets of the animal are marked with the orange lines.
Extended Data Fig. 5 Identification of ipRGCs on the MEA allow comparison of photoresponses at the population level.
Related to Fig. 4a. Schematic of the strategy used to record dark-adapted light responses from ipRGCs on the MEA. Photoreceptor-driven responses are initially recorded blindly from all RGCs (left). Intrinsic light sensitivity under synaptic blockade is then used to identify ipRGCs (middle). Finally, ipRGCs are clustered based on their light responses, and their rod- and cone-mediated photoresponses are analyzed retroactively (right).Recordings were performed as follows: Mice were dark-adapted; retinas were excised under infrared illumination to prevent light exposure; photoresponses from ipRGCs and RGCs were recorded blindly in regular Ames’ solution; synaptic blockers were then added, and light responses were recorded again.Analysis was conducted by first localizing all photoresponses to individual RGCs; then, intrinsic photoresponses under synaptic blockade were used to identify ipRGCs; finally, photoresponses of these ipRGCs were retroactively examined from recordings made prior to synaptic blockade. b. Intrinsic photoresponses of M1 ipRGCs (top) and non-M1 ipRGCs (bottom) to 20 sec blue (480 nm) illumination at increasing light intensities. c. M1 ipRGCs (n = 19 cells) exhibit high intrinsic photosensitivity compared to non-M1 ipRGCs (n = 50 cells). The error bar represents SEM. d. M1 ipRGCs exhibit faster time to peak compared to non-M1 ipRGCs. e. M1 ipRGCs exhibit an initial burst in spiking followed by suppression of spiking at high light intensities (13 log photons cm−2 s−1), consistent with depolarization block. n = 19 M1 & 50 non-M1ipRGCs, 4 retinas across 3 animals. f-k ipRGC photoresponses are suppressed under green light but not under violet light at high intensity. f. Illustration of dark adapted ipRGCs recorded on the MEA in response to long wavelength (green, λmax =535 nm) and short wavelength (violet, λmax =405 nm) increments in light intensity. g. Change in spike rate of dark adapted M1 ipRGCs (n=19 cells) in response to 3 sec visual stimuli from darkness across of range of green and violet light intensities. The error bar represents SEM. h. Population data showing peak spike rate for M1 ipRGCs (n=19 cells) across intensities. The error bar represents SEM. ** p = 0.0015, two-way ANOVA, Sidak’s multiple comparisons test. i. Sustained spike rate in M1 ipRGCs (n=19 cells) is suppressed at high intensity under green light but not under violet light. Plotted data shows mice as individual points with mean ± SEM. ** p = 0.0013, ns: not statistically different, two-way ANOVA post hoc Tukey. j. Same as in h, but for non-M1 ipRGCs (n=50 cells). The error bar represents SEM. ** p = 0.0084, two-way ANOVA, Sidak’s multiple comparisons test. k. Sustained spike rate in non-M1 ipRGCs (n=50 cells) is comparable at low and high intensities for both LEDs. Plotted data shows mice as individual points with mean ± SEM. ns is not statistically different, two-way ANOVA post hoc Tukey.
Extended Data Fig. 6
a-c. Actograms for wild type mice exposed to 15 min of sunlight and M1 ipRGC spike responses to sunlight and blue sky light. Related to Fig. 5. a. The double-plotted actograms of all wild type mice (n = 6). On day 8 the mice received a 15-min sunlight pulse (LP) at CT6. b. Average spike rates across cells in response to white light (n = 7 cells). SEM is indicated by colored shaded regions. Yellow vertical dashed line indicates the intensity of sunlight during midday. c. Average spike rates across cells in response to blue light (n = 13 cells). SEM is indicated by colored shaded regions. Blue vertical dashed line indicates the intensity of blue-sky during midday. Quantifications of observed phase shifts in a are shown in Fig. 5. d-f. Spectral properties of opsins and light sources used in the study. d. Normalized absorption spectra of S-opsin, Melanopsin, Rhodopsin and M-opsin. e. Normalized emission spectra of LED (375/385nm, 405 nm, 470 nm, 535 nm, white), sunlight and blue sky used in the study. f. Normalized S-opsin, Melanopsin, Rhodopsin and M-opsin content for each light source based on spectral overlap. g. Relative melanopsin activation across light sources.
Extended Data Fig. 7 Models for M1 ipRGC-SCN acts as an integrated pacemaker across the circadian cycle.
The diagram shows that ipRGCs send photic information to the SCN. Light phase-shifts the SCN only during the nighttime, but not during the daytime. The inability of light to shift the SCN clock during the daytime is attributed to the intrinsic property of the SCN clock in what is known as the “gate.” Here we demonstrate that an intrinsic property of ipRGCs, i.e., the depolarization block, also functions as a “gate” at the retina to confine the photic input to the SCN. This retinal gate acts in concert with the SCN gate to prevent phase shifts during the daytime. This retinal gate also limits the magnitude of phase shifts during nighttime when the SCN gate is open. Thus, we posit that the M1 ipRGC-SCN acts as an integrated pacemaker across the circadian cycle, with the retinal gate acting as a homeostatic gate, whereas the SCN gate acting as a circadian gate. a. During the daytime, the SCN “gate” is closed, and the ipRGC depolarization block characteristic, i.e., the retinal homeostatic gate, reduces the total number of ipRGCs that can send photic information to the SCN at defined intensities of light. This limited photic input cannot open the SCN “gate” to phase-shift the clock. b. During the nighttime, the SCN “gate” is open, and thus the photic input from ipRGCs can phase-shift the clock. However, the retinal homeostatic gate allows only limited photic input to the SCN to phase-shift the clock with a confined magnitude. c. During the daytime, chemogenetic activation of ipRGCs increases the total number of ipRGCs that sends retinal inputs to the SCN. This ample retinal input can force open the SCN “gate” to induce robust phase shift. Violet light, in compared to blue light or white light or even sunlight, also increases the total number of ipRGCs that sends the retinal input to the SCN. This increased retinal input can partially open the SCN “gate” to induce small magnitude of phase shift. d. During the nighttime, the SCN “gate” is open, and chemogenetic activation of ipRGCs increases the number of ipRGCs providing retinal input to the SCN. This ample retinal input induces robust phase shift.
Supplementary information
41586_2025_9894_MOESM1_ESM.docx
This file contains detailed methods and analyses for the following components: Pupillary Light Response; MEA Recordings and Validation of the New sOpn4flpO Mouse Line
Rights and permissions
About this article
Cite this article
Komal, R., Beier, C., Nath, A. et al. ipRGC properties prevent light from shifting the SCN clock during daytime. Nature (2026). https://doi.org/10.1038/s41586-025-09894-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-09894-z