Abstract
Rewriting RNA information to alter function requires controllable tools to edit RNA sequences within a user-defined region. Here we report a single-strand deaminase-assisted platform for adjustable RNA information manipulation (AIM). AIM is composed of a loop-forming guide RNA bound to an RNA-targeting Cas protein and an evolved TadA. AIM induces a loop, flanked by paired regions, in the target RNA; the loop size can be adjusted to allow conversions of single and multiple bases. We evolve TadA to achieve A-to-I, C-to-U or simultaneous A+C editing in coding and noncoding regions. We apply AIM to suppress the ochre nonsense codon (UAA) in disease-relevant cell and animal models, in which the two As must be simultaneously edited to rewrite the coding information. Moreover, we use AIM to manipulate adjacent phosphorylation sites important for protein function. Collectively, AIM is a versatile platform for manipulating RNA information within user-defined regions, opening additional avenues for functional RNA modulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequence data generated in this study were deposited to the National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE230140. Source data are provided with this paper.
References
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Porto, E. M., Komor, A. C., Slaymaker, I. M. & Yeo, G. W. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19, 839–859 (2020).
Pfeiffer, L. S. & Stafforst, T. Precision RNA base editing with engineered and endogenous effectors. Nat. Biotechnol. 41, 1526–1542 (2023).
Song, J., Zhuang, Y. & Yi, C. Programmable RNA base editing via targeted modifications. Nat. Chem. Biol. 20, 277–290 (2024).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
Rauch, S. et al. Programmable RNA-guided RNA effector proteins built from human parts. Cell 178, 122–134 (2019).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
Qian, Y. et al. Programmable RNA sensing for cell monitoring and manipulation. Nature 610, 713–721 (2022).
Jiang, K. et al. Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR. Nat. Biotechnol. 41, 698–707 (2022).
Kaseniit, K. E. et al. Modular, programmable RNA sensing using ADAR editing in living cells. Nat. Biotechnol. 41, 482–487 (2022).
Reautschnig, P. et al. CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat. Biotechnol. 40, 759–768 (2022).
Ojha, N., Diaz Quiroz, J. F. & Rosenthal, J. J. C. In vitro and in cellula site-directed RNA editing using the λNDD-BoxB system. Methods Enzymol. 658, 335–358 (2021).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Huang, X. et al. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 39, e104741 (2020).
Latifi, N., Mack, A. M., Tellioglu, I., Di Giorgio, S. & Stafforst, T. Precise and efficient C-to-U RNA base editing with SNAP-CDAR-S. Nucleic Acids Res. 51, e84 (2023).
Bhakta, S., Sakari, M. & Tsukahara, T. RNA editing of BFP, a point mutant of GFP, using artificial APOBEC1 deaminase to restore the genetic code. Sci. Rep. 10, 17304 (2020).
Han, W. et al. Programmable RNA base editing with a single gRNA-free enzyme. Nucleic Acids Res. 50, 9580–9595 (2022).
Stroppel, A. S. et al. Harnessing self-labeling enzymes for selective and concurrent A-to-I and C-to-U RNA base editing. Nucleic Acids Res. 49, e95 (2021).
Liu, Z., Jillette, N., Robson, P. & Cheng, A. W. Simultaneous multifunctional transcriptome engineering by CRISPR RNA scaffold. Nucleic Acids Res. 51, e77 (2023).
Song, J. et al. CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons. Mol. Cell 83, 139–155 (2023).
Adachi, H. et al. Targeted pseudouridylation: an approach for suppressing nonsense mutations in disease genes. Mol. Cell 83, 637–651 (2023).
Luo, N. et al. Near-cognate tRNAs increase the efficiency and precision of pseudouridine-mediated readthrough of premature termination codons. Nat. Biotechnol. 43, 114–123 (2025).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Tahmasebi, S., Khoutorsky, A., Mathews, M. B. & Sonenberg, N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 19, 791–807 (2018).
Sun, H. X., Li, K., Liu, C. & Yi, C. Q. Regulation and functions of non-m6A mRNA modifications. Nat. Rev. Mol. Cell Biol. 24, 714–731 (2023).
Fu, X. D. Non-coding RNA: a new frontier in regulatory biology. Natl Sci. Rev. 1, 190–204 (2014).
Ha, M. & Kim, V. N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014).
Xiang, J. S., Schafer, D. M., Rothamel, K. L. & Yeo, G. W. Decoding protein–RNA interactions using CLIP-based methodologies. Nat. Rev. Genet. 25, 879–895 (2024).
Bass, B. L. & Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 (1988).
Wagner, R. W., Smith, J. E., Cooperman, B. S. & Nishikura, K. A double-stranded-RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian-cells and Xenopus eggs. Proc. Natl Acad. Sci. USA 86, 2647–2651 (1989).
Melcher, T. et al. A mammalian RNA editing enzyme. Nature 379, 460–464 (1996).
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 41, 578–594 (2016).
Pecori, R., Di Giorgio, S., Lorenzo, J. P. & Papavasiliou, F. N. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat. Rev. Genet. 23, 505–518 (2022).
Wolf, J., Gerber, A. P. & Keller, W. tadA, an essential tRNA-specific adenosine deaminase from. EMBO J. 21, 3841–3851 (2002).
Losey, H. C., Ruthenburg, A. J. & Verdine, G. L. Crystal structure of tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159 (2006).
Yang, L. H. et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 7, 13330 (2016).
Lam, D. K. et al. Improved cytosine base editors generated from TadA variants. Nat. Biotechnol. 41, 686–697 (2023).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Lapinaite, A. et al. DNA capture by a CRISPR–Cas9-guided adenine base editor. Science 369, 566–571 (2020).
Rees, H. A., Wilson, C., Doman, J. L. & Liu, D. R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 5, eaax5717 (2019).
Li, J. A. et al. Structure-guided engineering of adenine base editor with minimized RNA off-targeting activity. Nat. Commun. 12, 2287 (2021).
Grunewald, J. et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 37, 1041–1048 (2019).
Zhou, C. Y. et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278 (2019).
Jeong, Y. K. et al. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 39, 1426–1433 (2021).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–U899 (2020).
Mort, M., Ivanov, D., Cooper, D. N. & Chuzhanova, N. A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 29, 1037–1047 (2008).
Stenson, P. D. et al. The Human Gene Mutation Database: 2008 update. Genome Med. 1, 13 (2009).
Bidou, L., Allamand, V., Rousset, J. P. & Namy, O. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 18, 679–688 (2012).
Bidou, L. et al. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 11, 619–627 (2004).
Martins-Dias, P. & Romao, L. Nonsense suppression therapies in human genetic diseases. Cell. Mol. Life Sci. 78, 4677–4701 (2021).
Welch, E. M. et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91 (2007).
Shi, N. et al. Restoration of dystrophin expression in mice by suppressing a nonsense mutation through the incorporation of unnatural amino acids. Nat. Biomed. Eng. 6, 195–206 (2022).
Lueck, J. D. et al. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 10, 822 (2019).
Albers, S. et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 618, 842–848 (2023).
Wang, J. M. et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 604, 343–348 (2022).
Wong, S. K., Sato, S. & Lazinski, D. W. Substrate recognition by ADAR1 and ADAR2. RNA 7, 846–858 (2001).
Matthews, M. M. et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 23, 426–433 (2016).
Azad, M. T. A., Bhakta, S. & Tsukahara, T. Site-directed RNA editing by adenosine deaminase acting on RNA for correction of the genetic code in gene therapy. Gene Ther. 24, 779–786 (2017).
Dugueperoux, I. et al. Cystic fibrosis at the Reunion Island (France): spectrum of mutations and genotype–phenotype for the Y122X mutation. J. Cyst. Fibros. 3, 185–188 (2004).
Karijolich, J. & Yu, Y. T. Therapeutic suppression of premature termination codons: mechanisms and clinical considerations (review). Int. J. Mol. Med. 34, 355–362 (2014).
Neugebauer, M. E. et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 41, 673–685 (2023).
Chen, L. et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat. Biotechnol. 41, 663–672 (2023).
Zhang, E., Neugebauer, M. E., Krasnow, N. A. & Liu, D. R. Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference. Nat. Commun. 15, 1697 (2024).
Li, Z. Y. et al. dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications. Nucleic Acids Res. 50, D471–D479 (2022).
Rao, R. S. P. & Moller, I. M. Large-scale analysis of phosphorylation site occupancy in eukaryotic proteins. Biochim. Biophys. Acta 1824, 405–412 (2012).
Schweiger, R. & Linial, M. Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol. Direct 5, 6 (2010).
Zheng, L. et al. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23, 1590–1601 (2003).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Yu, Y. et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 11, 2052 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Zhang, X. H. et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 38, 856–U810 (2020).
Li, C. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–U866 (2020).
Grünewald, J. et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 38, 861–U827 (2020).
Sakata, R. C. et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 38, 865–U846 (2020).
Weber, L. et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci. Adv. 6, eaay9392 (2020).
Antoniou, P. et al. Base-editing-mediated dissection of a γ-globin-regulatory element for the therapeutic reactivation of fetal hemoglobin expression. Nat. Commun. 13, 6618 (2022).
Lebek, S. et al. Ablation of CaMKIId oxidation by CRISPR–Cas9 base editing as a therapy for cardiac disease. Science 379, 179–185 (2023).
Yan, H. & Tang, W. Programmed RNA editing with an evolved bacterial adenosine deaminase. Nat. Chem. Biol. 20, 1361–1370 (2024).
Giudice, G., Sanchez-Cabo, F., Torroja, C. & Lara-Pezzi, E. ATtRACT—a database of RNA-binding proteins and associated motifs. Database (Oxf.) 2016, baw035 (2016).
Kluesner, M. G. et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Cheng, E. C. K., Lam, J. K. C. & Kwon, S. C. Cytosolic CRISPR RNAs for efficient application of RNA-targeting CRISPR–Cas systems. EMBO Rep. 26, 1891–1912 (2025).
Lu, B. et al. Transposase assisted tagmentation of RNA/DNA hybrid duplexes. Elife 9, e54919 (2020).
Lu, B. & Yi, C. TRACE-seq: rapid, low-input, one-tube RNA-seq library construction based on tagmentation of RNA/DNA hybrids. Curr. Protoc. 3, e735 (2023).
Acknowledgements
We thank the Optical Imaging Core Facility, National Center for Protein Sciences at Peking University for assistance with immunofluorescence experiments. We are grateful to Y. Luo, C. Shan and W. Gao for their help with sample preparation and data collection and to Y. Zhao (Indica Labs) for support with data analysis. We thank the National Center for Protein Sciences at Peking University, particularly G. Li and X. Zhang, for technical support with MGISEQ-2000 sequencing platforms and the Agilent 4150 TapeStation system. We also thank the Center for Quantitative Biology at Peking University for assistance with the ImageXpress Micro4 high-content imaging system and X. Li for help with data collection and analysis. We thank N. Luo and Q. Huang for sharing plasmids, Z. Wei for providing mouse serum and B. Lu, H. Sun and M. Zhang for the discussion. The data analysis was performed on the High-Performance Computing Platform at the School of Life Sciences, Peking University. This work was supported by the National Natural Science Foundation of China (22425071 and 22337001 to C.Y.), the Beijing Municipal Science and Technology Commission (Z231100002723005 to C.Y.) and the Ministry of Agriculture and Rural Affairs of China (NK2022010102 to C.Y.). This work was supported by the New Cornerstone Science Foundation through the XPLORER prize.
Author information
Authors and Affiliations
Contributions
C.Y. and Y.Z. conceptualized the project. Y.Z., Q.Z. and X.L. designed and performed the experiments with the assistance of Y.Y., P.G., R.Y., R.S., Y.Z. and A.W. H.W. analyzed all the sequencing data. Z.L., H.M., M.C. and H.X. participated in the design and interpretation of key experiments. C.Y., Y.Z., Q.Z., H.W. and X.L. wrote the manuscript. All the authors commented on and approved the paper.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed by Peking University for the technology disclosed in this publication. C.Y. and Y.Z. are coinventors on a patent application describing the AIM system. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
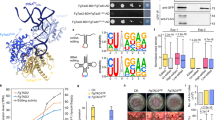
Extended Data Fig. 1 Identification of TadA variants for programmable RNA editing and investigation of optimal parameters for LF-gRNA design.
a, Structure superposition of tRNA anticodon loop in SaTadA-tRNA complex (PDB: 2B3J) and U-turn-shaped ssDNA substrate in ABE8e-sgRNA-DNA complex (PDB: 6VPC). b, Editing efficiencies of TadA variant fusion proteins with different LF-gRNAs. TadA variants include wild-type TadA (TadA-WT), TadA7.10, and TadA8e. c, Top: the representative on-target sequence in Reporter-UAG and the corresponding LF-gRNAs with diverse base-pairing arm lengths (ranging from 24 nt to 36 nt). Bottom: editing efficiencies of TadA7.10 and TadA8e fusion proteins with the aforementioned LF-gRNAs. n = 2 biological replicates. Data are presented as mean ± SD. d, Top: the representative on-target sequence in Reporter-UAG and the corresponding LF-gRNAs with non-symmetrical base-pairing arms. Bottom: editing efficiencies of TadA7.10 and TadA8e fusion proteins with different LF-gRNAs. n = 2 biological replicates. Data are presented as mean ± SD. e-f, Top: the representative on-target sequence in Reporter-UAG and the corresponding LF-gRNAs with different 3′ or 5′ arm length. Bottom: editing efficiencies of TadA8e fusion proteins with progressively shorten complementary 3′ arm (e) 5′ arm (f) and of the LF-gRNA.
Extended Data Fig. 2 Performance of AIM-A tools across different sequence contexts and different cell types.
a, Heatmap of 5′ and 3′ base preferences of AIM-Amax for all possible three-base motifs. b, Left: Representative on-target sequences of reporter constructs containing 6–10 consecutive adenosines. The LF-gRNA was used to induce loop sizes ranging from 6–10 nt on reporter transcripts. Right: Heatmap presenting editing efficiencies mediated by AIM-Amax. c. Schematic illustration showing the AIM editing window across LF-gRNA designs with varying loop sizes. d, EGFP + /mCherry+ ratio demonstrating the A-to-I editing activity of AIM-A and AIM-Amax across multiple cell lines. Cells were co-transfected with Reporter-UAG and AIM-A tools. e, Quantifying A-to-I editing rates on two endogenous transcripts mediated by AIM-Amax in N2A cells. f, Workflow to examine RNA editing rate, GAG content and Idua enzymatic activity in primary MEF cells from a Hurler syndrome mouse model containing homozygous Idua-W392X mutation (TGG-to-TAG). g, Quantification of A-to-I editing rates mediated by AIM-Amax in primary MEF cells. n = 3 independent biological replicates. Data are presented as mean ± SD. h, Histograms showing on-target and off-target A-to-I editing of the mouse Idua transcript using the AIM-Amax in primary MEF cells. Editing rates were quantified using NGS. i, The GAG content accumulation in WT, Idua-W392X and AIM-Amax-treated Idua-W392X primary MEF cells. Data represent mean values ± s.d. of n = 3 independently biological replicates. The P value was calculated by a two-tailed Student’s t-test. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. j, Measurement of Idua enzymatic activity in WT, Idua-W392X and AIM-Amax-treated Idua-W392X primary MEF cells. Data represent mean values ± s.d. of n = 3 independently biological replicates. The P value was calculated by a two-tailed Student’s t-test. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Extended Data Fig. 3 Suppression of ochre nonsense codon in disease-relevant cells and a mouse model.
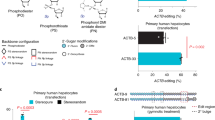
a, Normalized current-voltage relationships of HEK293T-CFTR-WT cells and HEK293T-CFTR-Y122X cells, showing total membrane currents at basal, stimulation, and inhibition states. HEK293T-CFTR-Y122X cells treated with CFTR-Y122X-targeting AIM-Amax were stimulated with 10 µM Forskolin and 10 µM IBMX, followed by perfusion with 10 µM CFTR inhibitor-172. HEK293T-CFTR-WT cells served as the positive control, while HEK293T-CFTR-Y122X cells treated with non-targeting AIM-Amax as the negative control. Data were collected from independent cells: n = 4. Data are presented as mean ± SEM. b, Schematics showing the structure of AAV constructs. c, In vivo RNA editing rates (UAA > UAG, UGA, and UGG) in AIM-Amax-treated DmdQ995X mice. Data represent mean values ± s.d. of n = 4 independent TA muscles. d, Western blot analysis of dystrophin protein expression in the TA muscles from WT, DmdQ995X, and AIM-Amax-treated DmdQ995X mice. e, H&E staining of TA muscles of WT, DmdQ995X, and AIM-Amax-treated DmdQ995X mice. Red arrows indicate inflammatory areas (shown in the wide view), and green arrows indicate normal myofibers without central nuclei (shown in the magnified view).
Extended Data Fig. 4 Performance of AIM-C tools across different sequence contexts and different cell types.
a, Heatmap of 5′ and 3′ base preferences of AIM-Cmax for all possible three-base motifs. b, EGFP + /mCherry+ ratio demonstrating the C-to-U editing activity of AIM-C and AIM-Cmax across multiple cell lines. Cells were co-transfected with Reporter-CAC and AIM-C tools. c, Quantifying C-to-U editing rates on two endogenous transcripts mediated by AIM-Cmax in N2A cells. d, Comparison of AIM-Cmax and existing RNA C-to-U editing tools (RESCUE, RESCUE-S, and CURE-N) at three transcripts. Data are presented as mean values (n = 2) and each dot represents an individual value.
Extended Data Fig. 5 Codon and amino acid changes enabled by AIM-A, AIM-C, and AIM-A&C.
a, Circos plots showing the specific codon changes enabled by AIM-A (left), AIM-C (middle), and AIM-A&C (right), respectively. AIM-A induces 61 different types of codon changes, 46 of which (orange, 75.4%) result in amino acid substitutions. AIM-C can induce 61 different types of codon change, with 43 (green, 70.5%) leading to amino acid changes. AIM-A&C can induce 30 different types of codon alterations, 29 of which (red, 97%) change amino acids. b, Tables summarizing the codon and amino acid changes enabled by AIM-A (left), AIM-C (middle), and AIM-A&C (right). Codon changes leading to different amino acids, induced by the AIM-A or AIM-C tools, are underlined in black. Codons uniquely altered by AIM-A&C are highlighted in green.
Extended Data Fig. 6 Manipulation of single or adjacent PTM sites.
a, Bar-plot showing the distribution of PTMs among various amino acid residues according to the dbPTM database. b, Quantification of concurrent editing rates at the second A of the Y340 codon and the second A of the Y341 codon in endogenous RAF1 transcripts using NGS. c, Quantification of concurrent editing rates at the second C of the S363 codon and the second C of the S364 codon in the endogenous BRAF transcripts using NGS. d, Quantification of concurrent editing rates at the second C of the first codon and the first A of the second codon in the endogenous CDK13 transcripts using NGS.
Extended Data Fig. 7 Transcriptome-wide specificity of AIM.
a, Sequence context motif plots of identified A-to-I and C-to-U off-targets, showing the preferred binding sequence context for the REPAIRv1, REPAIRv2, AIM-A, AIM-Amax and AIM-A&Cmax systems. b, Jitter plot showing the transcriptome-wide A-to-I off-target events in WT, AIM-Amax-treated, and untreated Idua mutant primary MEF cells. AIM-Amax was delivered via AAV-DJ. c, Scatter plots showing the comparison of the global gene expression levels between control and AIM-treated primary MEF cells. Pearson’s correlation coefficient analysis was used to assess the differential gene expressions.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12 and Tables 1–9.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhuang, Y., Zhu, Q., Wu, H. et al. Single-strand deaminase-assisted editing for functional RNA manipulation. Nat Biotechnol (2026). https://doi.org/10.1038/s41587-025-02956-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02956-7