Abstract
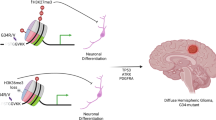
Primary mismatch-repair-deficient high-grade gliomas (priMMRD-HGG) are lethal tumors characterized by hypermutation, resistance to chemoradiation and variable response to immunotherapy. To investigate the mechanisms governing the emergence of driver mutations and their impact on gliomagenesis and patient outcomes, we analyzed genomic and clinical data from 162 priMMRD-HGG. Here we identified three subgroups defined by secondary driver mutations in replicative DNA polymerases or IDH1. These subgroups converge on glioma drivers through distinct combinations of genomic instability–generating mechanisms, displaying an inverse correlation between point mutations and copy number alterations. MMRD signatures drive the emergence of specific mutations in TP53 and IDH1, notably excluding common pediatric glioma drivers. Global hypomethylation stratifies priMMRD-HGG into a unique methylation cluster. DNA-polymerasemut priMMRD-HGG exhibit ultrahypermutation, an immune-hot microenvironment and immunotherapy responsiveness, whereas IDH1mut priMMRD-HGG are immune-cold and immunotherapy resistant. MMRD-driven gliomagenesis defines the role of nonrandom mutagenesis patterns in cancer development, providing frameworks for targeted and immune-therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Methylation array data for this cohort are available through Gene Expression Omnibus (GEO) at GSE307219. Previously published and newly generated WES data for matched tumor and normal pairs are available in the repository in anonymized format (EGAD50000001794). These data are under controlled access to protect the identities of research participants and will be available by application to the Replication Repair Deficiency Data Access Committee. Data access requests take approximately 2–3 weeks to review. NanoString expression data were previously published41 and are available through GEO at GSE227756. The remaining processed clinical and molecular characterization data are available within Supplementary Data 1. Material requests are available from U.T., including a relevant research proposal to be approved by the IRRDC Governance Committee (replicationrepair.ca).
Code availability
No custom code or software packages were developed as part of this study. Use of existing software packages and versions is indicated with their respective methods.
References
Negrini, S., Gorgoulis, V. G. & Halazonetis, T. D. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010).
Loeb, L. A. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer 11, 450–457 (2011).
Tomasetti, C., Li, L. & Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017).
Preston, B. D., Albertson, T. M. & Herr, A. J. DNA replication fidelity and cancer. Semin. Cancer Biol. 20, 281–293 (2010).
Shlien, A. et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 47, 257–262 (2015).
Tabori, U. et al. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin. Cancer Res. 23, e32–e37 (2017).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Schindler, G. et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 121, 397–405 (2011).
Wu, G. et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 46, 444–450 (2014).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Das, A., Ercan, A. B. & Tabori, U. An update on central nervous system tumors in germline replication-repair deficiency syndromes. Neurooncol. Adv. 6, vdae102 (2024).
Negm, L. et al. The landscape of primary mismatch repair deficient gliomas in children, adolescents, and young adults: a multi-cohort study. Lancet Oncol. 26, 123–135 (2025).
Amayiri, N. et al. High frequency of mismatch repair deficiency among pediatric high grade gliomas in Jordan. Int. J. Cancer 138, 380–385 (2016).
Touat, M. et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 580, 517–523 (2020).
Tauziède-Espariat, A. et al. Refinement of diagnostic criteria for pediatric-type diffuse high-grade glioma, IDH- and H3-wildtype, MYCN-subtype including histopathology, TP53, MYCN and ID2 status. Acta Neuropathol. Commun. 11, 170 (2023).
Suwala, A. K. et al. Primary mismatch repair deficient IDH-mutant astrocytoma (PMMRDIA) is a distinct type with a poor prognosis. Acta Neuropathol. 141, 85–100 (2021).
Das, A. et al. Genomic predictors of response to PD-1 inhibition in children with germline DNA replication repair deficiency. Nat. Med. 28, 125–135 (2022).
Ercan, A. B. et al. Clinical and biological landscape of constitutional mismatch-repair deficiency syndrome: an International Replication Repair Deficiency Consortium cohort study. Lancet Oncol. 25, 668–682 (2024).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Chung, J. et al. DNA polymerase and mismatch repair exert distinct microsatellite instability signatures in normal and malignant human cells. Cancer Discov. 11, 1176–1191 (2021).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016).
Petralia, F. et al. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell 183, 1962–1985 (2020).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Holmfeldt, L. et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45, 242–252 (2013).
Shlien, A. et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc. Natl Acad. Sci. USA 105, 11264–11269 (2008).
Hoelscher, M. et al. SELDI-TOF analysis of glioblastoma cyst fluid is an approach for assessing cellular protein expression. Neurol. Res. 35, 993–1001 (2013).
Hassel, B., Niehusmann, P., Halvorsen, B. & Dahlberg, D. Pro-inflammatory cytokines in cystic glioblastoma: a quantitative study with a comparison with bacterial brain abscesses. With an MRI investigation of displacement and destruction of the brain tissue surrounding a glioblastoma. Front. Oncol. 12, 846674 (2022).
De Leeuw, B. I., Van Baarsen, K. M., Snijders, T. J. & Robe, P. A. J. T. Interrelationships between molecular subtype, anatomical location, and extent of resection in diffuse glioma: a systematic review and meta-analysis. Neurooncol. Adv. 1, vdz032 (2019).
Louis, D. N. et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 23, 1231 (2021).
Suwala, A. K. et al. Glioblastomas with primitive neuronal component harbor a distinct methylation and copy-number profile with inactivation of TP53, PTEN, and RB1. Acta Neuropathol. 142, 179–189 (2021).
Dodgshun, A. J. et al. Germline-driven replication repair-deficient high-grade gliomas exhibit unique hypomethylation patterns. Acta Neuropathol. 140, 765–776 (2020).
Hadad, S. et al. De novo replication repair deficient glioblastoma, IDH-wildtype’ is a distinct glioblastoma subtype in adults that may benefit from immune checkpoint blockade. Acta Neuropathol. 147, 3 (2023).
Turcan, S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Poulos, R. C., Olivier, J. & Wong, J. W. H. The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 45, 7786–7795 (2017).
Németh, E. et al. Two main mutational processes operate in the absence of DNA mismatch repair. DNA Repair (Amst.) 89, 102827 (2020).
Tomkova, M. et al. Human DNA polymerase ε is a source of C>T mutations at CpG dinucleotides. Nat. Genet. 56, 2506–2516 (2024).
Campbell, B. B. et al. Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056 (2017).
Fang, H. et al. Deficiency of replication-independent DNA mismatch repair drives a 5-methylcytosine deamination mutational signature in cancer. Sci. Adv. 7, eabg4398 (2021).
Flynn, A., Waszak, S. M. & Weischenfeldt, J. Somatic CpG hypermutation is associated with mismatch repair deficiency in cancer. Mol. Syst. Biol. 20, 1006–1024 (2024).
Levine, A. B. et al. Immuno-oncologic profiling of pediatric brain tumors reveals major clinical significance of the tumor immune microenvironment. Nat. Commun. 15, 5790 (2024).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018).
Zhang, X. et al. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro Oncol. 18, 1402–1412 (2016).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Kohanbash, G. et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J. Clin. Invest. 127, 1425–1437 (2017).
Durno, C. et al. Survival benefit for individuals with constitutional mismatch repair deficiency undergoing surveillance. J. Clin. Oncol. 39, 2779–2790 (2021).
Ahadova, A. et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int. J. Cancer 143, 139–150 (2018).
Sekine, S. et al. Mismatch repair deficiency commonly precedes adenoma formation in Lynch syndrome-associated colorectal tumorigenesis. Mod. Pathol. 30, 1144–1151 (2017).
Bolivar, A. M. et al. Genomic landscape of lynch syndrome colorectal neoplasia identifies shared mutated neoantigens for immunoprevention. Gastroenterology 166, 787–801 (2024).
Fang, H. et al. Mutational processes of distinct POLE exonuclease domain mutants drive an enrichment of a specific TP53 mutation in colorectal cancer. PLoS Genet. 16, e1008572 (2020).
Bai, R., Lv, Z., Xu, D. & Cui, J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 8, 34 (2020).
Larkin, T. et al. Upfront adjuvant immunotherapy of replication repair-deficient pediatric glioblastoma with chemoradiation-sparing approach. JCO Precis. Oncol. 5, 1426–1431 (2021).
Das, A. et al. Combined immunotherapy improves outcome for replication-repair-deficient (RRD) high-grade glioma failing anti–PD-1 monotherapy: a report from the international RRD consortium. Cancer Discov. 14, 258–273 (2024).
Ahmad, O., Ahmad, T. & Pfister, S. M. IDH mutation, glioma immunogenicity, and therapeutic challenge of primary mismatch repair deficient IDH-mutant astrocytoma PMMRDIA: a systematic review. Mol. Oncol. 18, 2822–2841 (2024).
Acknowledgements
U.T. was supported by the Canadian Institutes for Health Research (CIHR) (PJT-156006), the CIHR Joint Canada-Israel Health Research Program (MOP-137899), a Stand Up to Cancer (SU2C)—Bristol Myers Squibb Catalyst Research (SU2C-AACR-CT07-17) grant and the Department of Defense Rare Cancers Research Program (HT9425-24-1-1096). This research is also generously supported by SickKids Foundation donors Harry and Agnieszka Hall, Meagan’s Hug (MW-2014-10), BRAINchild Canada and The LivWise Foundation. U.T. and G.G. were supported by a Children’s Oncology Group National Cancer Institute Community Oncology Research Program Research Base Administrative Supplement Request (3UG1CA189955-08S2). U.T., C.H., P.B.D. and A.L. were supported by a Terry Fox Research Institute Program Projects Grant at The Hospital for Sick Children (TFRI Project 1156-03). P.B.D. was supported by the Canadian Institutes for Health Research (PJT180605). A.D. was supported by the Cannonball Kids’ Cancer Foundation Young Investigator Grant in Partnership with Kindred Foundation and a Rally Foundation for Childhood Cancer Research award (25CDN08). D.M. is supported in part by the CIBC Children’s Foundation Chair in Child Health Research. Y.E.M. is supported by an ISF grant (2794/21). A.S. is supported by the Garron Family Chair in Childhood Cancer Research and the Canadian Institutes for Health Research (CIHR). G.G. was partially funded by the Paul C. Zamecnik Chair in Oncology at the Mass General Cancer Center. The other authors declare no relevant funding sources.
Author information
Authors and Affiliations
Contributions
N.R.F., A.D. and U.T. conceived of and designed the research. N.R.F., Y.C. and J.R.D. performed the processing of WES data. N.R.F., A.L. and C.L. performed the immunohistochemistry and RNA NanoString analyses. A.R. and B.E.-W. assessed the magnetic resonance imaging. L. Negm, J.C. and Y.E.M. performed the low-pass whole-genome sequencing and MSI analyses. A.B.E., L.S., V.B., M.E., S.D., L. Nobre, J.B., A.V., D.M., V.R., A.H., E.B., M.A., P.B.D., A.S. and C.H. collected the samples and provided tissue annotations and clinicopathological information. N.R.F., O.A., A.J.D., D.T.W.J., S.M.P., A.D. and U.T. performed the methylation array analyses and interpretation. J.M.H. and G.G. provided the allele-specific copy number profiling. N.R.F., N.M.N., A.D. and U.T. interpreted the data. N.R.F., N.M.N., A.D. and U.T. wrote and edited the paper. U.T. supervised the study. All authors approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
A.S. is a cofounder of, and holds equity in, NewCode Oncology, which has licensed technology from The Hospital for Sick Children (SickKids). C.H. discloses honorarium from Servier. D.T.W.J. is a founder of and stock shareholder in Heidelberg Epignostix GmbH. E.B. discloses participation in an advisory board for Servier. G.G. receives research funds from IBM, Pharmacyclics/AbbVie, Bayer, Genentech, Calico, Ultima Genomics, Inocras, Google, Kite and Novartis, and is also an inventor on patent applications filed by the Broad Institute related to MSMuTect, MSMutSig, POLYSOLVER, SignatureAnalyzer-GPU, MSEye, MinimuMM-seq and DLBclass. He is a founder, consultant and holds privately held equity in Scorpion Therapeutics; is a founder of, and holds privately held equity in, Predicta Biosciences; and holds privately held equity in Antares Therapeutics. J.B. discloses participation in advisory boards for Servier Canada and Alexion-Canada. J.M.H. is an employee of and holds privately held equity in Predicta Biosciences. S.M.P. discloses honoraria from BioSkryb, waived honoraria from Epignostix GmbH, PMC Advisory Board and University Hospital Essen Westdeutsches Tumorzentrum, unpaid memberships in GPOH, SIOP, DGNN, DKG, AACR and SNO, and is a stock shareholder in Epignostix GmbH. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks David Capper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characteristics of the study cohort.
Flowchart of the study cohort obtained from the International Replication Repair Deficiency Consortium (IRRDC). LOGIC, low-coverage genomic instability characterization.
Extended Data Fig. 2 Characterization of mutational signatures and driver events in priMMRD glioma.
a, Average Cosmic v3.2 SBS signature contributions across priMMRD subgroups. Signatures are colored and grouped by etiology. b, Total microsatellite InDel burden across priMMRD-HGG subgroups. Colored diamonds indicate median value. Two-sided Wilcoxon rank-sum test with Benjamini–Hochberg correction. Colored diamonds represent the median value. c, Polymerase epsilon (POLE) deficiency-specific microsatellite instability signature scores across priMMRD subgroups21. Two-sided Wilcoxon rank-sum test with Benjamini–Hochberg correction. Colored diamonds represent the median value. d, Oncoprint of glioma driver mutations and copy number alterations according to priMMRD-HGG subgroups and previously published cohorts of pediatric and adult MMRP glioma22,23. HGG, high-grade glioma; LGG, low-grade glioma; codel, 1p/19q co-deleted.
Extended Data Fig. 3 Copy number variation profiles of priMMRD glioma.
a, Copy number alteration proportions across priMMRD subgroups. Selected genes commonly affected by copy number variation in glioma are indicated. b, Percentage of genome altered between subgroups given as the percentage of bases affected by copy number gains or losses over the total number of covered bases. Two-sided Wilcoxon rank-sum test with Benjamini–Hochberg correction. Colored diamonds represent the median value.
Extended Data Fig. 4 Mechanisms of mutagenesis define priMMRD subgroups.
a, Frequencies of alteration types in recurrent glioma drivers across priMMRD subgroups. Two-sided Fisher’s exact test with Benjamini–Hochberg correction. b, Allele-specific copy number profiles of near-haploid priMMRD-2 gliomas. c, Somatic point mutations of glioma drivers found in haploid gliomas. d, Age at diagnosis in years across near-haploid (n = 3) and diploid (n = 155) priMMRD-HGG. Two-sided Wilcoxon rank-sum test. e, Tumor mutation burden (TMB); (SNVs per megabase) across near-haploid (n = 3) and diploid (n = 159) priMMRD-HGG. Two-sided Wilcoxon rank-sum test. Colored diamonds represent the median value in d and e.
Extended Data Fig. 5 Classification of priMMRD glioma methylation profiles.
a, Heidelberg Brain Tumor Classifier v12.8 classes of priMMRD-HGG (n = 82) subgroups. No classification indicates samples with a confidence score below 0.9. b, Uniform manifold approximation and projection (UMAP) plot of priMMRD-HGG and reference classes from the Heidelberg Brain Tumor Classifier v12.8 and primary mismatch repair-deficient IDH-mutant astrocytoma (PMMRDIA). PriMMRD-HGG subgroups are indicated by outlined shapes according to the included key. Samples are colored according to their v12.8 class prediction or lack of classification (prediction score <0.9). Samples that failed classification but clustered with PMMRDIA are colored in blue.
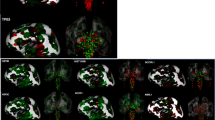
Extended Data Fig. 6 Characteristics of priMMRD glioma epigenetic outliers.
a, Table of priMMRD-HGG that fall outside of the main priMMRD methylation cluster on t-SNE with a proposed reason for their clustering. b, Representative H3K27me3 immunohistochemistry (IHC) staining of the two priMMRD-HGG that clustered with the DMG_K27 methylation class (n = 1, respectively). Only tumor 4 had confirmed H3-3A K27M and H2K27me3 loss, suggesting that tumor 5 may have noncanonical mechanisms of chromatic landscape alteration. c, Estimated cumulative distribution of average β values plotted individually by priMMRD-HGG. The three priMMRD-HGG that clustered with DMG_K27 or DHG_G34 are indicated. d, Average β values of the three priMMRD-HGG that clustered with DMG_K27 or DHG_G34 versus those that did not. Colored diamonds represent the median value. e, Histogram of variant allele frequency (VAF) distributions in tumor 4, VAF of H3-3A p.K27M is indicated, demonstrating a clonal mutation. f, Histogram of variant allele frequency (VAF) distributions in tumor 6, VAF of H3-3A p.G34R is indicated, demonstrating a clonal mutation. g, Copy number profile of tumor 6 with copy gain of chromosome 1q, which includes H3-3A. h, Copy number profile of tumor 7 with MYCN amplification. CRTL_CORPCAL, control tissue corpus callosum; DHG_G34, diffuse hemispheric glioma H3.3 G34 mutant; DMG_K27, diffuse midline glioma H3 K27M; GBM_MYCN, glioblastoma subclass MYCN; HGG_E, high-grade glioma subtype E; IHG, infantile hemispheric glioma; PA_INF, pilocytic astrocytoma subclass infratentorial; pedHGG_MYCN, pediatric high-grade glioma subclass MYCN; pedHGG_RTK1A, pediatric high-grade glioma subclass RTK1A.
Extended Data Fig. 7 priMMRD glioma harbors a distinct spectrum of TP53 hotspot mutations.
a, Mutational spectrum of TP53 hotspot mutations in priMMRD-HGG subgroups as well as previously published IDH-mutant and IDH-wildtype glioma cohorts10,22. b, Recurrently mutated codons in TP53 across priMMRD and MMRP glioma subtypes. For codons with multiple protein changes, they are listed in order of frequency. c, Average mutational spectra of priMMRD-HGG and hotspot mutations in ATRX, NF1 and RB1 driven by MMRD signatures.
Extended Data Fig. 8 SBS96D has high similarity to time signatures and MMRd-A.
From top to bottom, mutational spectra of the MutSα-specific de novo mutational signature SBS96D, mutational spectra of SBS96D reconstructed from COSMIC v3.2 SBS signature loadings. Cosine similarity between SBS96D and the reconstructed profile is given, mutational spectra of the COSMIC v.3.2 SBS signatures that form the components of SBS96D and mutational spectra of previously published MutSα-specific mutational signature MMRd-A36,39.
Extended Data Fig. 9 Extended survival analyses of priMMRD glioma.
a, Overall survival of priMMRD-HGG by subgroup, which did not receive immune checkpoint inhibitor therapy. b, Forest plot of the Cox-proportional hazards model of priMMRD-1 glioma treated with either anti-PD1 monotherapy or chemoirradiation. c, Forest plot of Cox-proportional hazards model of priMMRD-2 glioma treated with either anti-PD1 monotherapy or chemoirradiation. d, Forest plot of the Cox-proportional hazards model of priMMRD-3 glioma treated with either anti-PD1 monotherapy or chemoirradiation. e, Forest plot of Cox-proportional hazards model of priMMRD-HGG treated with anti-PD1 monotherapy, adjusted for subgroup, MMRD syndrome, age and sex. Error bars indicate a 95% confidence interval for b through e.
Supplementary information
Supplementary Data 1
Clinical and methylation classification data of the priMMRD glioma cohort. Clinical, methylation, expression and genomic data and metadata of the priMMRD glioma cohort.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fernandez, N.R., Chang, Y., Nunes, N.M. et al. Patterns of hypermutation shape tumorigenesis and immunotherapy response in mismatch-repair-deficient glioma. Nat Genet 58, 132–142 (2026). https://doi.org/10.1038/s41588-025-02420-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02420-x