Abstract
During recent years, the use of libraries-scale genomic manipulations scaffolded on CRISPR guide RNAs have been transformative. However, these existing approaches are typically multiplexed across genomes. Unfortunately, building cells with multiple, nonadjacent precise mutations remains a laborious cycle of editing, isolating an edited cell and editing again. The use of bacterial retrons can overcome this limitation. Retrons are genetic systems composed of a reverse transcriptase and a noncoding RNA that contains an multicopy single-stranded DNA, which is reverse transcribed to produce multiple copies of single-stranded DNA. Here we describe a technology—termed a multitron—for precisely modifying multiple sites on a single genome simultaneously using retron arrays, in which multiple donor-encoding DNAs are produced from a single transcript. The multitron architecture is compatible with both recombineering in prokaryotic cells and CRISPR editing in eukaryotic cells. We demonstrate applications for this approach in molecular recording, genetic element minimization and metabolic engineering.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and its Supplementary Information or will be made available from the authors upon request. Sequencing data associated with this study are available on NCBI SRA as BioProject ID PRJNA1107632.
Code availability
Custom code to process or analyze data from this study is available via GitHub at https://github.com/Shipman-Lab/multitrons (https://doi.org/10.5281/zenodo.11289190).
References
Shalem, O., Sanjana, N. E. & Zhang, F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 16, 299–311 (2015).
Doench, J. G. Am I ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet. 19, 67–80 (2018).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Nyerges, Á. et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc. Natl Acad. Sci. USA 115, E5726–E5735 (2018).
Barbieri, E. M., Muir, P., Akhuetie-Oni, B. O., Yellman, C. M. & Isaacs, F. J. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171, 1453–1467.e13 (2017).
Isaacs, F. J. et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353 (2011).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Tong, Y. et al. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc. Natl Acad. Sci. USA 116, 20366–20375 (2019).
Tong, Y., Jørgensen, T. S., Whitford, C. M., Weber, T. & Lee, S. Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 12, 5206 (2021).
Volke, D. C., Martino, R. A., Kozaeva, E., Smania, A. M. & Nikel, P. I. Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing. Nat. Commun. 13, 3026 (2022).
Yuan, Q. & Gao, X. Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat. Commun. 13, 2771 (2022).
Aulicino, F. et al. Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus. Nucleic Acids Res. 50, 7783–7799 (2022).
Li, H. et al. Multiplex precision gene editing by a surrogate prime editor in rice. Mol. Plant 15, 1077–1080 (2022).
Gao, L. et al. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084 (2020).
Millman, A. et al. Bacterial retrons function in anti-phage defense. Cell 183, 1551–1561.e12 (2020).
Mestre, M. R., González-Delgado, A., Gutiérrez-Rus, L. I., Martínez-Abarca, F. & Toro, N. Systematic prediction of genes functionally associated with bacterial retrons and classification of the encoded tripartite systems. Nucleic Acids Res. 48, 12632–12647 (2020).
Bobonis, J. et al. Bacterial retrons encode phage-defending tripartite toxin–antitoxin systems. Nature 609, 144–150 (2022).
Yee, T., Furuichi, T., Inouye, S. & Inouye, M. Multicopy single-stranded DNA isolated from a Gram-negative bacterium, Myxococcus xanthus. Cell 38, 203–209 (1984).
Inouye, S., Hsu, M. Y., Eagle, S. & Inouye, M. Reverse transcriptase associated with the biosynthesis of the branched RNA-linked msDNA in Myxococcus xanthus. Cell 56, 709–717 (1989).
Lampson, B. C., Inouye, M. & Inouye, S. Reverse transcriptase with concomitant ribonuclease H activity in the cell-free synthesis of branched RNA-linked msDNA of Myxococcus xanthus. Cell 56, 701–707 (1989).
Simon, A. J., Ellington, A. D. & Finkelstein, I. J. Retrons and their applications in genome engineering. Nucleic Acids Res. 47, 11007–11019 (2019).
Farzadfard, F. & Lu, T. K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014).
Sharon, E. et al. Functional genetic variants revealed by massively parallel precise genome editing. Cell 175, 544–557.e16 (2018).
Simon, A. J., Morrow, B. R. & Ellington, A. D. Retroelement-based genome editing and evolution. ACS Synth. Biol. 7, 2600–2611 (2018).
Schubert, M. G. et al. High-throughput functional variant screens via in vivo production of single-stranded DNA. Proc. Natl Acad. Sci. USA 118, e2018181118 (2021).
Lopez, S. C., Crawford, K. D., Lear, S. K., Bhattarai-Kline, S. & Shipman, S. L. Precise genome editing across kingdoms of life using retron-derived DNA. Nat. Chem. Biol. 18, 199–206 (2022).
Jiang, W. et al. High-efficiency retron-mediated single-stranded DNA production in plants. Synth. Biol. 7, ysac025 (2022).
Fishman, C. B. et al. Continuous multiplexed phage genome editing using recombitrons. Preprint at bioRxiv https://doi.org/10.1101/2023.03.24.534024 (2023)
Mosberg, J. A., Lajoie, M. J. & Church, G. M. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics 186, 791–799 (2010).
Wannier, T. M. et al. Improved bacterial recombineering by parallelized protein discovery. Proc. Natl Acad. Sci. USA 117, 13689–13698 (2020).
Palka, C., Fishman, C. B., Bhattarai-Kline, S., Myers, S. A. & Shipman, S. L. Retron reverse transcriptase termination and phage defense are dependent on host RNase H1. Nucleic Acids Res. 50, 3490–3504 (2022).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Ellington, A. J. & Reisch, C. R. Efficient and iterative retron-mediated in vivo recombineering in Escherichia coli. Synth Biol (Oxf) 7, ysac007 (2022).
Alper, H., Jin, Y. S., Moxley, J. F. & Stephanopoulos, G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7, 155–164 (2005).
Kang, M. J. et al. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol. Bioeng. 91, 636–642 (2005).
Jin, Y. S. & Stephanopoulos, G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 9, 337–347 (2007).
Chen, H., Bjerknes, M., Kumar, R. & Jay, E. Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22, 4953–4957 (1994).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Cunningham, F. X. J., Sun, Z., Chamovitz, D., Hirschberg, J. & Gantt, E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell 6, 1107–1121 (1994).
Liu, W. et al. Retron-mediated multiplex genome editing and continuous evolution in Escherichia coli. Nucleic Acids Res. 51, 8293–8307 (2023).
DiCarlo, J. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR–Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
DiCarlo, J. et al. Yeast oligo-mediated genome engineering (YOGE). ACS Synth. Biol. 2, 741–749 (2013).
Roy, K. et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat. Biotechnol. 36, 512–520 (2018).
Guo, X. et al. High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR–Cas9 in yeast. Nat. Biotechnol. 36, 540–546 (2018).
Świat, M. et al. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 45, 12585–12598 (2017).
Ferreira, R. et al. Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae. ACS Synth. Biol. 7, 10–15 (2018).
Liang, Z. et al. Advanced eMAGE for highly efficient combinatorial editing of a stable genome. Preprint at bioRxiv https://doi.org/10.1101/2020.08.30.256743 (2020).
Niwa, H., Yamamura, K. & Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991).
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Acknowledgements
Work was supported by funding from the National Science Foundation (MCB 2137692), the National Institute of Biomedical Imaging and Bioengineering (R21EB031393), the National Institute of General Medical Sciences (1DP2GM140917) and the UCSF Program for Breakthrough Biomedical Research. S.L.S. is a Chan Zuckerberg Biohub—San Francisco investigator and acknowledges additional funding support from the L.K. Whittier Foundation and the Pew Biomedical Scholars Program. A.G.-D. was supported by the California Institute of Regenerative Medicine (CIRM) scholar program. S.C.L. was supported by a Berkeley Fellowship for Graduate Study.
Author information
Authors and Affiliations
Contributions
S.L.S., A.G.-D. and S.C.L. conceived the study. A.G.-D., S.C.L. and M.R.-M. cloned plasmids used in this study, A.G.-D. performed all experiments with E. coli, and S.C.L. performed all experiments with S. cerevisiae and human cultured cells. M.R.M. and C.B.F. performed NGS and prepped and ran the sequencing libraries. A.G.-D., S.C.L. and S.L.S. designed the work, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.G.-D., S.C.L. and S.L.S. are named inventors on a patent application related to the technologies described in this work (63/524,317). S.L.S. is a founder of Retronix Bio. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
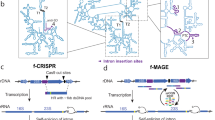
Extended Data Fig. 1 Trans msr multitron architecture enables precise genome editing.
Top: Schematic of retron recombineering using an msd array with a single msr sequence in trans including a terminator (T) between the msd array and msr. Bottom: quantification of precise editing rates for precise editing of rpoB or gyrA simultaneously by Illumina sequencing after 24 h of editing. Circles show each of the three biological replicates, bars are mean ± SD.
Extended Data Fig. 2 Optimization of retron recombineering using a single plasmid.
a. Left: schematic of the different retron operon architectures tested. ncRNA with donor (orange and blue), genes required (grey) and optimized ribosome binding sites (RBS) regions (green) are indicated Right: quantification of rates for precise rpoB editing, circles show each of the three biological replicates, bars are mean ± SD. b. Quantification of precise editing rates for rpoB target site at 30 and 37 °C, circles show each of the three biological replicates, lines are mean ± SD. c. Quantification of OD600 using increasing concentrations of m-toluic acid after 16 h of bacterial growing (n = 1). d. Quantification of precise editing rates for rpoB using different concentrations of arabinose (n = 1). e. Quantification of colonies with intact msd arrays. A total of 30 colonies coming from 3 different replicates were sequenced, bars are mean ± SD All precise editing rates were quantified using Illumina MiSeq after 24 h of editing. f. Scheme of the protocol used to analyze genetic stability of the retron arrays. Briefly, recombineering plasmid was transformed into E. coli strain bMS.346, followed by 5 days of growing and diluting in the presence or absence of the arabinose. A dilution of the final culture was diluted and plated. Finally, the msd Array of 10 individual colonies per replicate (n = 3) were amplified and sequenced to assess genetic stability of the multitron approach.
Extended Data Fig. 3 Local off-target mutations.
a. Quantification of precise editing rates for fbaH and hda genes using a live or dead version of Eco1 RT, circles show each of the three biological replicates, bars are mean ± SD. b. Local off-target mutation frequency in the 70 bp region of the chromosome homologous to fbaH and hda editing donors using a live of dead version of Eco1 RT circles show each of the three biological replicates, bars are mean ± SD. All data was quantified using Illumina MiSeq after 24 h of editing.
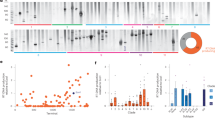
Extended Data Fig. 4 Intended and undesired on-target mutation rates caused by arrayed retron multiplexed editing in yeast cells.
a. Top: Schematic of the donor encoding retron ncRNA/gRNA expression cassette expressed from a Gal7 Pol II promoter and flanked by ribozymes versus a new construction replacing ribozymes with Csy4 sequences. Bottom left: schematic of a retron ncRNA-Cas9 gRNA hybrid for genome editing in yeast, depicted above the protein-coding expression cassette which is inserted into the yeast genome. Bottom right: quantification of indel rates of the ADE2 locus in yeast by Illumina sequencing after 48 h of editing. Circles show each of the three biological replicates, bars are mean ± SD; absence/presence of Csy4 in the protein-coding expression cassette is shown below the graph. b. Top: schematic of an arrayed retron ncRNA-Cas9 gRNA expression cassette, expressed from a Gal7 Pol II promoter, flanked by ribozymes, and separated by a Csy4 sequence. The retron editors in positions 1 and 2 target the ADE2 and FAA1 locus, respectively. Bottom: quantification of indel rates of the ADE2 and FAA1 loci in yeast by Illumina sequencing after 48 h of editing. Circles show each of the three biological replicates, bars are mean ± SD; absence/presence of Csy4 in the protein-coding expression cassette is shown below the graph. c. Top: schematic of an arrayed retron msdRNA-Cas9 gRNA expression cassette, expressed from a Gal7 Pol II promoter, flanked and separated by a Csy4 sequence; the msrRNA is expressed in trans from a SNR52 Pol III promoter. Bottom: assembly schematic for one-pot Golden Gate cloning of multiple msdRNA-sgRNA editors. d. Schematic showing the presumed processing, annealing and reverse-transcription involved in the generation of editing donors from arrayed retron msdRNA-Cas9 gRNA cassettes. e. top: schematic of 5x arrayed retron msdRNA-Cas9 gRNA expression cassettes, as shown in Extended Data Fig. 4c. Bottom: quantification of precise editing of the various yeast loci targeted by the retron editors shown above, by Illumina sequencing, after 24 and 120 h of editing. The editors target ADE2, CAN1, TRP2, SGS1 and FAA1. Two-way ANOVA, effect of expression time, P = 0.0038. Circles show each 3 biological replicates, bars are mean ± SD. f–h, top: schematic of 2x, 3x or 5x arrayed retron msdRNA-Cas9 gRNA expression cassettes. Bottom: quantification of indel rates of the various yeast loci targeted by the retron editors shown above, by Illumina sequencing, after 24 and 120 h of editing. Individual open circles show each of three biological replicates per condition, bars are mean ± SD The editors target ADE2 and FAA1 (f); ADE2, CAN1 and FAA1 (g); and ADE2, CAN1, TRP2, SGS1 and FAA1 (h).
Supplementary information
Supplementary Information
Supplementary Tables 1–6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
González-Delgado, A., Lopez, S.C., Rojas-Montero, M. et al. Simultaneous multi-site editing of individual genomes using retron arrays. Nat Chem Biol 20, 1482–1492 (2024). https://doi.org/10.1038/s41589-024-01665-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41589-024-01665-7
This article is cited by
-
Construction of synthetic protein-binding non-genetic DNA systems in living cells
Nature Chemistry (2026)
-
Discovery and engineering of retrons for precise genome editing
Nature Biotechnology (2025)
-
An experimental census of retrons for DNA production and genome editing
Nature Biotechnology (2025)
-
Structural basis of the RNA-mediated Retron-Eco2 oligomerization
Cell Discovery (2025)
-
Reducing competition between msd and genomic DNA improves retron editing efficiency
EMBO Reports (2024)