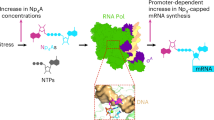
Abstract
Enigmatic dinucleoside tetraphosphates, known as ‘alarmones’ (Np4Ns), have recently been shown to function in bacteria as precursors to Np4 caps on transcripts, likely influencing RNA longevity and cellular adaptation to stress. In proteobacteria, ApaH is the predominant enzyme that hydrolyzes Np4Ns and decaps Np4-capped RNAs to initiate their 5′-end-dependent degradation. Here we conducted a biochemical and structural study to uncover the catalytic mechanism of Escherichia coli ApaH, a prototypic symmetric Np4N hydrolase, on various Np4Ns and Np4-capped RNAs. We found that the enzyme uses a unique combination of nonspecific and semispecific substrate recognition, enabling substrates to bind in two orientations with a slight orientational preference. Despite such exceptional recognition properties, ApaH efficiently decaps various Np4-capped mRNAs and sRNAs, thereby impacting their lifetimes. Our findings highlight the need to determine substrate orientation preferences before designing substrate-mimicking drugs, as enzymes may escape activity modulation with one of the alternative substrate orientations.

This is a preview of subscription content, access via your institution
Access options






Similar content being viewed by others
Data availability
Coordinates of the structures were deposited in the Protein Data Bank with PDB ID codes: 9OJD, apo ApaH; 9OJP, apo ApaH with Mn2+; 9OJX, cocrystallized with GDP; 9OK1, cocrystallized with CDP; 9ON7, cocrystallized with ppAG; 9OLN, cocrystallized with NAD+; 9OND, cocrystallized with ppAGG; 9OJQ, soaked ADP, active conformation; 9OJW, soaked ADP, inactive conformation; 9OK2, soaked UDP; 9OLZ, soaked Ap4A; 9OM9, soaked AppCH2ppA; 9OMC, soaked Gp4G; 9OMU, soaked Gp4A; 9OMW, soaked Up4A; 9OMX, soaked Up4U; 9ON0, D37A variant, soaked Up4A; 9OLY, apo D37A variant; 9ONG, soaked Ap4AG; 9OQ9, soaked Ap4AGG; 9OON, soaked Ap4GU; 9OOY, soaked Gp4AU; 9OPH, soaked Up4AG; 9OQB, soaked Up4AGG; 9OPG, soaked Ap4UG; 9OP2, soaked Ap4GUAA. The MD simulation results were deposited in Zenodo (https://doi.org/10.5281/zenodo.15594844 (ref. 77)). Source data are provided with this paper.
References
Finamore, F. J. & Warner, A. H. The occurrence of P1, P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J. Biol. Chem. 238, 344–348 (1963).
Zamecnik, P. C., Stephenson, M. L., Janeway, C. M. & Randerath, K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 91–97 (1966).
Plateau, P. & Blanquet, S. Dinucleoside oligophosphates in micro-organisms. Adv. Micro. Physiol. 36, 81–109 (1994).
Coste, H., Brevet, A., Plateau, P. & Blanquet, S. Non-adenylylated bis(5′-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 262, 12096–12103 (1987).
Lee, P. C., Bochner, B. R. & Ames, B. N. AppppA, heat-shock stress, and cell oxidation. Proc. Natl Acad. Sci. USA 80, 7496–7500 (1983).
Varshavsky, A. Diadenosine 5′, 5″′-P1, P4-tetraphosphate: a pleiotropically acting alarmone? Cell 34, 711–712 (1983).
Bochner, B. R., Lee, P. C., Wilson, S. W., Cutler, C. W. & Ames, B. N. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37, 225–232 (1984).
Ferguson, F., McLennan, A. G., Urbaniak, M. D., Jones, N. J. & Copeland, N. A. Re-evaluation of diadenosine tetraphosphate (Ap4A) from a stress metabolite to bona fide secondary messenger. Front. Mol. Biosci. 7, 606807 (2020).
Lee, P. C., Bochner, B. R. & Ames, B. N. Diadenosine 5′,5″′-P1,P4-tetraphosphate and related adenylylated nucleotides in Salmonella typhimurium. J. Biol. Chem. 258, 6827–6834 (1983).
Giammarinaro, P. I. et al. Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis. Nat. Microbiol. 7, 1442–1452 (2022).
Lee, Y. N., Nechushtan, H., Figov, N. & Razin, E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity 20, 145–151 (2004).
Luciano, D. J., Levenson-Palmer, R. & Belasco, J. G. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell 75, 957–966 e8 (2019).
Hudecek, O. et al. Dinucleoside polyphosphates act as 5′-RNA caps in bacteria. Nat. Commun. 11, 1052 (2020).
Frantisek Potuznik, J. et al. Diadenosine tetraphosphate (Ap(4) A) serves as a 5′ RNA cap in mammalian cells. Angew. Chem. Int. Ed. Engl. 63, e202314951 (2024).
Randerath, K., Janeway, C. M., Stephenson, M. L. & Zamecnik, P. C. Isolation and characterization of dinucleoside tetra- and tri-phosphates formed in the presence of lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 98–105 (1966).
Luciano, D. J. & Belasco, J. G. Np4A alarmones function in bacteria as precursors to RNA caps. Proc. Natl Acad. Sci. USA 117, 3560–3567 (2020).
Benoni, R., Culka, M., Hudecek, O., Gahurova, L. & Cahova, H. Dinucleoside polyphosphates as RNA building blocks with pairing ability in transcription initiation. ACS Chem. Biol. 15, 1765–1772 (2020).
Mititelu, M. B. et al. Arabidopsis thaliana NudiXes have RNA-decapping activity. RSC Chem. Biol. 4, 223–228 (2023).
Levenson-Palmer, R. et al. A distinct RNA recognition mechanism governs Np4 decapping by RppH. Proc. Natl Acad. Sci. USA 119, e2117318119 (2022).
Plateau, P., Fromant, M., Brevet, A., Gesquiere, A. & Blanquet, S. Catabolism of bis(5′-Nucleosidyl) oligophosphates in Escherichia coli: metal requirements and substrate-specificity of homogeneous diadenosine-5′,5″′-P1,P4-tetraphosphate pyrophosphohydrolase. Biochemistry 24, 914–922 (1985).
Farr, S. B., Arnosti, D. N., Chamberlin, M. J. & Ames, B. N. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl Acad. Sci. USA 86, 5010–5014 (1989).
Leveque, F., Blanchin-Roland, S., Fayat, G., Plateau, P. & Blanquet, S. Design and characterization of Escherichia coli mutants devoid of Ap4N-hydrolase activity. J. Mol. Biol. 212, 319–329 (1990).
Johnstone, D. B. & Farr, S. B. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10, 3897–3904 (1991).
Nishimura, A. et al. Diadenosine 5′,5″′-P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 2, 401–413 (1997).
Ismail, T. M., Hart, C. A. & McLennan, A. G. Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar Typhimurium to invade cultured mammalian cells. J. Biol. Chem. 278, 32602–32607 (2003).
Hansen, S., Lewis, K. & Vulic, M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52, 2718–2726 (2008).
Monds, R. D. et al. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J. Bacteriol. 192, 3011–3023 (2010).
Kimura, Y., Tanaka, C., Sasaki, K. & Sasaki, M. High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology 163, 86–93 (2017).
Ji, X. et al. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Natl Acad. Sci. USA 116, 9578–9585 (2019).
Wang, Q. H., Hu, W. X., Gao, W. & Bi, R. C. Crystal structure of the diadenosine tetraphosphate hydrolase from Shigella flexneri 2a. Proteins 65, 1032–1035 (2006).
Almo, S. C. et al. Structural genomics of protein phosphatases. J. Struct. Funct. Genomics 8, 121–140 (2007).
Jin, Y. et al. Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia. Structure 32, 1197–1207 (2024).
Minazzato, G. et al. Functional characterization of COG1713 (YqeK) as a novel diadenosine tetraphosphate hydrolase family. J. Bacteriol. 202, e00053-20 (2020).
Guranowski, A., Jakubowski, H. & Holler, E. Catabolism of diadenosine 5′,5″′-P1,P4-tetraphosphate in procaryotes. Purification and properties of diadenosine 5′,5″′-P1,P4-tetraphosphate (symmetrical) pyrophosphohydrolase from Escherichia coli K12. J. Biol. Chem. 258, 4784–4789 (1983).
Martin, J. E., Waters, L. S., Storz, G. & Imlay, J. A. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 11, e1004977 (2015).
Sasaki, M., Takegawa, K. & Kimura, Y. Enzymatic characteristics of an ApaH-like phosphatase, PrpA, and a diadenosine tetraphosphate hydrolase, ApaH, from Myxococcus xanthus. FEBS Lett. 588, 3395–3402 (2014).
Cahova, H., Winz, M. L., Hofer, K., Nubel, G. & Jaschke, A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015).
Kowtoniuk, W. E., Shen, Y., Heemstra, J. M., Agarwal, I. & Liu, D. R. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl Acad. Sci. USA 106, 7768–7773 (2009).
Chen, Y. G., Kowtoniuk, W. E., Agarwal, I., Shen, Y. & Liu, D. R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009).
Wang, J. et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 47, e130 (2019).
Bailey, S. et al. The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms. Structure 10, 589–600 (2002).
Guranowski, A., Brown, P., Ashton, P. A. & Blackburn, G. M. Regiospecificity of the hydrolysis of diadenosine polyphosphates catalyzed by three specific pyrophosphohydrolases. Biochemistry 33, 235–240 (1994).
Doamekpor, S. K., Sharma, S., Kiledjian, M. & Tong, L. Recent insights into noncanonical 5′ capping and decapping of RNA. J. Biol. Chem. 298, 102171 (2022).
Jiang, Y. L. et al. Structural and enzymatic characterization of the streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH. J. Biol. Chem. 286, 35906–35914 (2011).
Barta, M. L., Lovell, S., Sinclair, A. N., Battaile, K. P. & Hefty, P. S. Chlamydia trachomatis CT771 (nudH) is an asymmetric Ap4A hydrolase. Biochemistry 53, 214–224 (2014).
Mancini, F. & Cahova, H. The mysterious world of non-canonical caps—what we know and why we need new sequencing techniques. ChemBioChem 26, e202400604 (2025).
Uhrig, R. G., Kerk, D. & Moorhead, G. B. Evolution of bacterial-like phosphoprotein phosphatases in photosynthetic eukaryotes features ancestral mitochondrial or archaeal origin and possible lateral gene transfer. Plant Physiol. 163, 1829–1843 (2013).
Castaneda Londono, P. A., Banholzer, N., Bannermann, B. & Kramer, S. Is mRNA decapping by ApaH like phosphatases present in eukaryotes beyond the Kinetoplastida? BMC Ecol. Evol. 21, 131 (2021).
Kramer, S. The ApaH-like phosphatase TbALPH1 is the major mRNA decapping enzyme of trypanosomes. PLoS Pathog. 13, e1006456 (2017).
Gao, A. et al. Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF. Nucleic Acids Res. 46, 6841–6856 (2018).
Windsor, I. W. & Raines, R. T. A substrate selected by phage display exhibits enhanced side-chain hydrogen bonding to HIV-1 protease. Acta Crystallogr. D Struct. Biol. 74, 690–694 (2018).
Marquez-Monino, M. A. et al. Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping. Sci. Adv. 7, eabf6744 (2021).
Cao, H., Pauff, J. M. & Hille, R. Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid. J. Biol. Chem. 285, 28044–28053 (2010).
Yadav, P. K. et al. S-3-carboxypropyl-l-cysteine specifically inhibits cystathionine γ-lyase-dependent hydrogen sulfide synthesis. J. Biol. Chem. 294, 11011–11022 (2019).
Coleman, T. M., Wang, G. & Huang, F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 32, e14 (2004).
Vasilyev, N. & Serganov, A. Preparation of short 5′-triphosphorylated oligoribonucleotides for crystallographic and biochemical studies. Methods Mol. Biol. 1320, 11–20 (2016).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Galindo-Murillo, R. et al. Assessing the current state of amber force field modifications for DNA. J. Chem. Theory Comput. 12, 4114–4127 (2016).
Zgarbova, M. et al. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 7, 2886–2902 (2011).
Mark, P. & Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960 (2001).
Bradbrook, G. M. et al. X-Ray and molecular dynamics studies of concanavalin-A glucoside and mannoside complexes relating structure to thermodynamics of binding. J. Chem. Soc. Faraday Trans. 94, 1603–1611 (1998).
Frisch, M. J. et al. Gaussian 16 https://gaussian.com/gaussian16/ (Gaussian Inc., 2016).
Case, D. et al. Amber 2021 http://ambermd.org (University of California, San Francisco, 2021).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Parrinello, M. & Rahman, A. Polymorphic transitions in single-crystals: a new molecular-dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A. & Moreno, E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291 (2021).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Nuthanakanti, A. et al. MD data for ‘ApaH decaps Np4N-capped RNAs in two alternative orientations’. Zenodo https://doi.org/10.5281/zenodo.15594844 (2025).
Acknowledgements
This research used the Northeastern Collaborative Access Team beamlines, funded by the NIH (P30 GM124165), at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under contract DE-AC02-06CH11357. This research also used beamlines 17-ID-1 (AMX) and 17-ID-2 (FMX) of the National Synchrotron Light Source II, a U.S. DOE Office of Science User Facility operated by Brookhaven National Laboratory under contract DE-SC0012704. Beamline operation is supported by the Center for BioMolecular Structure, funded by the NIH (P30 GM133893) and the DOE Office of Biological and Environmental Research (FWP BO070). R.S.B. is a Damon Runyon Dale F. Frey awardee supported by the Damon Runyon Cancer Research Foundation (DRG-50-22). This research was also supported by the Hirschfelder Professorship Fund and the Research Forward Fund from the University of Wisconsin–Madison (to X.H.) and by the NIH (grants R35GM145359 and R01GM035769 to J.G.B., R37CA289040 to R.S.B. and R01GM112940 and R21GM151508 to A.S.).
Author information
Authors and Affiliations
Contributions
A.N. determined the X-ray structures and conducted biochemical experiments. M.K. and N.R.B. performed MS experiments under the supervision of R.S.B. R.L.-P. conducted decapping experiments with long RNAs under the supervision of J.G.B. Y.W. and X.H. conceived and carried out MD simulations. J.G.B. and A.S. wrote the manuscript with the help of A.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Ping Yin and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Specificity of Np4N hydrolysis and RNA decapping by E. coli ApaH.
(a,b) Representative chromatograms showing the kinetics of Ap4A (a) and Ap4G (b) hydrolysis. (c) Sequences and expected secondary structures of long RNA substrates. Left, a representative RNA substrate bearing two RNA hairpins. RNAs used in the study shared the same stem–loop structures. Right, Ap4A8XL RNA used as an internal standard. This RNA contains three RNA hairpins and a long unpaired 5′ segment. (d) A representative gel showing the kinetics of Ap4G0 RNA decapping. Ap4G0 was mixed with the internal standard Ap4A8XL and treated with ApaH. Decapping of each RNA was monitored as a function of time by boronate gel electrophoresis and fluorescence. (e) Decapping of the invariant internal standard Ap4A8XL (Std). Left to right, decapping of Ap4A8XL in the reactions graphed in Fig. 2a–c, respectively. Time points and error bars represent mean ± s.d. of three independent measurements (n = 3). (f–h) Inhibition of ApaH by reaction products. IC50 values, calculated by fitting the data to the ‘inhibitor vs. response’ model of GraphPad Prism, are indicated for Ap4A + ppAG (f) and Ap4AGG + ppAGG (h). Data points and error bars are mean ± s.d. of three independent measurements (n = 3) for Ap4A+ppAG. Data points for all other reactions are from 2 independent measurements (n = 2). The reactions proceeded for 5 min. (f) Inhibition of Np4A hydrolysis by ppAG, ADP, and UDP. (g) Inhibition of Ap4AG hydrolysis by ADP. (h) Inhibition of Ap4AGG hydrolysis by ppAGG.
Extended Data Fig. 2 Details of cation and NDP recognition by ApaH.
Close-up views of the Mn2+ cations in the apo structure with supplemented Mn2+ (a), soaked with Mg2+ (b), and soaked with ADP (c). Red, violet, and green spheres represent water molecules, Mn2+ and Mg2+ cations, respectively. Final refined structures are shown with the unbiased simulated annealing composite omit maps (light blue mesh) contoured at 1.0–1.1σ levels. Magenta mesh represents the anomalous map at 4 (a) and 3 (b,c) σ levels. (d–g) Unbiased maps (0.8–1.0σ levels) for ApaH-bound ADP (d), GDP (e), CDP (f), and UDP (g). Amino acids contacting the adenine of ADP are in sticks. Dashed dark blue and red lines represent putative hydrogen bonds and CH–π interactions. (h) Superposition of the ADP-bound catalytically competent (gray) and incompetent (light orange) conformations. Top: the active site showing amino acids interacting with phosphates in the catalytically incompetent conformation. A yellow dashed line connects water molecule W1 with the β phosphorus atom. Magenta ovals indicate new or changed hydrogen bonds. Bottom, an arrangement involving the reactive water molecule, Mn2+ cations, and the β-phosphate. Blue arrows show different positions of W1 and the β-phosphate in the two structures. The distance between W1 and the phosphorus atom and the angles between W1, the phosphorus atom, and the non-bridging oxygen atoms are depicted in yellow. (i) Superposition of the apo (cyan) and ADP-bound (gray) structures. Red spheres represent water molecules in the apo structure, including Mn2+-coordinated waters and an extra water molecule (extra W) found in the place of the α-phosphate. Blue arrows show shifts upon ADP binding. (j) ADP binding improves the quality of the unbiased maps (1.0σ level). Trp249 from the apo and ADP-bound structures is in cyan and gray colors, respectively.
Extended Data Fig. 3 Structural and biochemical studies of ApaH targeting Np4N substrates and their analogs.
(a) Hydrolysis of 0.1 mM Ap4A and Ap4AG by 70 nM ApaH in 1 mM NaF for 5 min. The bars are means of two independent experiments (n = 2). (b) Hydrolysis of 0.5 mM AppCH2ppA (AppcppA) by 0.3 mM ApaH in 4 mM Mg2+ and 10 mM Ca2+ for 3 h. (c) Hydrolysis of 0.5 mM NAD+ by 0.3 mM ApaH in 4 mM Mg2+ and 10 mM Ca2+ for 3 h. (d,e) The active site bound to Ap4A (d) and Up4A (e). Refined structures are shown with unbiased simulated annealing composite omit maps (light blue mesh) contoured at 1.0σ level. The adenosine (magenta) and four phosphates are visible in the map and were built into the structure, while the second nucleoside (yellow) was only modeled to show the feasibility of fitting it into the structure. (f) Replacement of Mn1 by a Mg2+ cation in the Ap4A-bound structure. Mn1 has weaker unbiased (top, light blue mesh, 6.0σ level) and anomalous (bottom, magenta mesh, 4.0σ level) maps than Mn2 and was modeled as a mixture of Mn2+ and Mg2+. (g–i) Views of the bound Up4U (g), Gp4G (h), and AppCH2ppA (i) shown with an unbiased map. (j) Superposition of the Np4N-bound structures. Gp4A, orange; Ap4A, magenta; Up4A, purple; Up4U, cyan; Gp4G, dark green; and AppCH2ppA, green. (k) A structural diagram of NAD+. The shading corresponds to the visible (ADP) and disordered (nicotinamide ribose (NR)) moieties. (l) The bound NAD+ with an unbiased map (0.9σ level). The model in yellow represents a visible map and may account for one of the possible positions of the moiety. (m) Superposition of the NAD+-bound (in colors) and ADP-bound (gray) structures.
Extended Data Fig. 4 Effects of mutations on the catalytic activity of ApaH.
(a) Conservation of ApaH in bacteria. The E. coli ApaH sequence is shown with secondary structure elements. Levels of strict sequence conservation were determined from a multiple sequence alignment of a representative set of moderately similar ApaH sequences, as described in Methods. Amino acid identities are: 100%, magenta; 90%, red; 80%, orange; 70%, yellow. The residues involved in metal coordination are indicated in green boxes. (b) Projection of amino acid conservation onto the Ap4A-bound structure. Color code as in (a). (c) Kinetics of Ap4A hydrolysis by ApaH mutants. Data points are from two independent experiments (n = 2) with similar results. The full-time course and initial time points fitted to an exponential decay model and used to calculate rate constants (indicated) are shown in the left and right panels, respectively. (d) A close-up view of the catalytic site of the Asp37Ala mutant in the apo state, shown with unbiased simulated annealing omit maps (light blue mesh) contoured at the 1.0σ level. The red dashed circle indicates the loss of the map for Mn2. (e) Superposition of the catalytic sites from the wild type (gray) and Asp37Ala mutant (colored) bound to Ap4U. Please note that two alternative conformations of the phosphate moieties in the mutant do not align well with the corresponding moieties in the wild-type protein, as shown by a blue arrow. (f–i) Hydrolysis of Ap4A by ApaH mutants: Glu232Ala (f), Glu202Ala (g), Trp249Ala and Trp249Phe (h), and Ser230Gly (i). Left, full-time course, and right, initial rates of hydrolysis. The decapping curve for the WT protein is shown as a dashed line. Data points are from two independent experiments (n = 2) with similar results.
Extended Data Fig. 5 Structural evidence for bidirectional binding of RNA to ApaH.
(a–f) Close-up views of the active site bound to the ppAG (a), Ap4AGG (b), Up4AG (c), Ap4GU (d), Up4AGG (e), and Ap4GUAA (f) RNAs. The refined structures are shown with a simulated annealing composite omit map (light blue mesh) contoured at 0.9–1.0σ level. Dashed circles highlight the areas corresponding to the phosphate of the +2 nucleotide or, in the ppAG structure (a), the area for the γ-phosphate and δ-phosphate of Np4N. Double-headed arrows indicate the orientation of the RNAs in the catalytic site. Arrowhead size is proportional to 18O labeling in the mass spectrometry experiments (Fig. 5c). Blue arrows point to the alternative conformations of the β-phosphate in the ppAG structure (a).
Extended Data Fig. 6 Conformational variability of the disordered RNA regions assessed by MD simulations of the RNA–ApaH complexes.
(a,b) Structural models of the Ap4AG (cyan and magenta)-bound ApaH (green) in the downward (a) and upward (b) orientations used for MD simulations (e). (c) The binding free energy of Ap4AG to ApaH in the downward (cyan) and upward (magenta) orientations. Each bar is a mean ± s.d. derived from ten bootstrapped ensembles (n = 10, using resampled trajectories with replacement). A p-value was calculated using a two-sided t-test based on the average binding free energies from the ten bootstrapped ensembles, with no adjustments for multiple comparisons. (d) Stability of RNA–ApaH binding in MD simulations (e). The plots show time-dependent distance changes between the center of mass of the tetraphosphate group in Ap4AG and the combined center of mass of residues R41 and R184. Results from five independent MD simulations, separated by gray dashed lines, for each system are concatenated. (e) Conformational variability of the disordered RNA regions. Two-dimensional heatmaps show conformational ensembles sampled from restrained MD simulations of the indicated ApaH-bound RNAs. The axes correspond to the first two principal components (PCs) derived from principal component analysis of the Cartesian coordinates of the flexible RNA regions. The adenosine of each RNA was positioned in the nucleoside binding site so that the RNAs with adenosine in the cap (0) or (+1) position were modeled in the downward and upward conformations. The Ap4 moiety was restrained during simulations. The colors indicate conformational density estimated via kernel density estimation, with darker colors representing more frequent conformations. (f) Root mean square fluctuation (RMSF) for each nucleotide of the RNA for the simulations from (e). Each bar is RMSF from 4,000 frames, with a saving interval of 1 ns between two consecutive frames. The systems with RNA in the downward and upward orientations are teal and purple.
Extended Data Fig. 7 The frequency of contacts between RNA and ApaH in MD simulations.
(a–f) The contact analysis was performed based on the simulations in Extended Data Fig. 6e,f for the following RNAs: (a) Ap4GU, (b) Gp4AU, (c) Ap4GUC, (d) Gp4AUC, (e) Ap4GUCU, and (f) Gp4AUCU. The contacts are defined by the distance smaller than 4 Å between heavy protein atoms of amino acid side chains and unrestrained parts of the RNAs. Ap4 moieties were restrained during the MD simulations. The plots (left panels) show the contact frequency, calculated as the ratio of frames with a contact to the total number of frames. For each simulated nucleotide of the RNA (0, +1, +2, +3 or +4), the top ten protein residues are listed. The images (right panels) visualize the contact frequencies on representative protein–RNA complex structures from the MD simulations. RNAs are in cyan sticks. Protein residues selected for mutagenesis are shown as sticks, with darker colors representing higher contact frequencies in each system. The contact residues also contributing to the interactions with the Ap4 group are in red in the left panels and are shown as red lines in the right panels. Mn²⁺ ions are shown as spheres.
Extended Data Fig. 8 Analysis of a positively charged surface near the active site of ApaH.
(a) Multiple sequence alignment for the region corresponding to amino acids 75–88 of E. coli ApaH. The entire protein sequences were aligned well, as evidenced by the absence of gaps and the perfect alignment of the flanking regions, which contain highly conserved residues such as Asn65, His66, Asp67, and Trp101. The figure shows representative sequences with various numbers and distributions of positively charged residues, as well as the absence of these residues in the 75–88 region. (b–e). Effects of the mutations on the hydrolysis of various substrates by E. coli ApaH. Hydrolysis of Ap4A (b,c), Ap4AG (c), and Up4AG (d). Left, full-time course and right, initial rates of hydrolysis, shown with the initial rate constants (k, s−1). The decapping curve for the WT protein is shown as a dashed line. Data points are from two independent experiments (n = 2) with similar results.
Supplementary information
Supplementary Information
Supplementary Tables 1–7 and Supplementary Figs. 1–5.
Source data
Source Data Fig. 2 and Extended Data Fig. 1
Unprocessed gel images for Fig. 2a–c and Extended Data Fig. 1e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nuthanakanti, A., Korn, M., Levenson-Palmer, R. et al. ApaH decaps Np4N-capped RNAs in two alternative orientations. Nat Chem Biol (2025). https://doi.org/10.1038/s41589-025-01991-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41589-025-01991-4