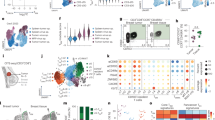
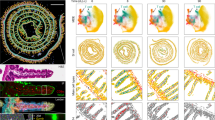
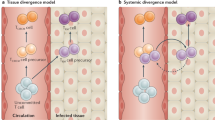
Abstract
The presence of CD8+ T cells coexpressing residency and exhaustion molecules in chronic diseases often correlate with clinical outcomes; however, the relationship between these cells and conventional tissue-resident memory (TRM) cells or exhausted CD8+ T (TEX) cells is unclear. Here we show that chronic antigen stimulation drives development of tissue-resident TEX (TR-TEX) cells that are distinct from TRM cells generated after antigen clearance. TR-TEX and TRM cells are regulated by different transcriptional networks with only TR-TEX cells being Tox-dependent for residency programming. While TEX cells (including TR-TEX) are unable to generate TRM cells after antigen withdrawal, TRM cells differentiate into TEX cells upon chronic antigen exposure. Cell-state-specific transcriptional signatures reveal a selective association of TR-TEX cells with patient responses to immune checkpoint blockade, and only TR-TEX but not TRM cells responded to PD-1 pathway inhibition in vivo. These data suggest that TR-TEX and TRM cells are developmentally divergent cell states that share a tissue-residency program but have distinct roles in disease control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Multimodal ADT, HTO, RNA and ATAC raw sequencing data generated have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE278342. Raw differential gene expression, differential accessibility data and unique gene signatures generated in this study are available as Supplementary Tables. Publicly available datasets analyzed during this manuscript were GSE70813, GSE179613 and GSE199565, or via gutcellatlas.org.
Code availability
No custom code was generated in this study. All bioinformatics analyses were performed using publicly available packages and code in the order and with the parameters and thresholds described in the Methods. Sequencing analysis outputs (for example DEGs and DACRs) are provided as Supplementary Tables.
Change history
13 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41590-026-02420-x
References
Christo, S. N., Park, S. L., Mueller, S. N. & Mackay, L. K. The multifaceted role of tissue-resident memory T cells. Annu. Rev. Immunol. 42, 317–345 (2024).
Szabo P. A., Miron M., Farber D. L. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. https://doi.org/10.1126/sciimmunol.aas9673 (2019).
Milner, J. J. et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Crowl, J. T. et al. Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol. 23, 1121–1131 (2022).
Mackay, L. K. et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Artic. Nat. Immunol. 14, 1294–1301 (2013).
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009).
Park, S. L. et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19, 183–191 (2018).
Schenkel, J. et al. cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014).
Park, S. L. et al. Tissue-resident memory CD8+ T cells promote melanoma–immune equilibrium in skin. Nature 565, 366–371 (2019).
Wijeyesinghe, S. et al. Expansible residence decentralizes immune homeostasis. Nature 592, 457–462 (2021).
McLane L., Abdel-Hakeem M., Wherry E. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. https://doi.org/10.1146/annurev-immunol-041015-055318 (2019).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165 (2016).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160 (2016).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Yao, C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019).
Lan, X., Zebley, C. C. & Youngblood, B. Cellular and molecular waypoints along the path of T cell exhaustion. Sci. Immunol. 8, eadg3868 (2023).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Christo, S. N. et al. Discrete tissue microenvironments instruct diversity in resident memory T cell function and plasticity. Nat. Immunol. 22, 1140–1151 (2021).
Isaacs, J. F. et al. CD39 is expressed on functional effector and tissue-resident memory CD8+ T cells. J. Immunol. 213, 588–599 (2024).
Kurd N. S. et al. Early precursors and molecular determinants of tissue-resident memory CD8(+) T lymphocytes revealed by single-cell RNA sequencing. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aaz6894 (2020).
Wherry, E. J., Blattman, J. N., Murali-Krishna, K., van der Most, R. & Ahmed, R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77, 4911–4927 (2003).
Im, S. J., Konieczny, B. T., Hudson, W. H., Masopust, D. & Ahmed, R. PD-1+ stemlike CD8 T cells are resident in lymphoid tissues during persistent LCMV infection. Proc. Natl Acad. Sci. USA 117, 4292 (2020).
Beura, L. K. et al. Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J. Leukoc. Biol. 97, 217–225 (2015).
Scott, M. C. et al. Deep profiling deconstructs features associated with memory CD8(+) T cell tissue residence. Immunity 58, 162–181.e10 (2025).
Beltra, J. C. et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841.e8 (2020).
Gavil, N. V. et al. Chronic antigen in solid tumors drives a distinct program of T cell residence. Sci. Immunol. 8, eadd5976 (2023).
Boland, B. S. et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 5, eabb4432 (2020).
Ryan, G. E., Harris, J. E. & Richmond, J. M. Resident memory T cells in autoimmune skin diseases. Front. Immunol. 12, 652191 (2021).
Gavil, N. V., Cheng, K. & Masopust, D. Resident memory T cells and cancer. Immunity 57, 1734–1751 (2024).
Boddupalli, C. S. et al. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight 1, e88955 (2016).
Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021).
Banchereau R. et al. Intratumoral CD103+ CD8+ T cells predict response to PD-L1 blockade. J. Immunother. Cancer. https://doi.org/10.1136/jitc-2020-002231 (2021).
Duhen, T. et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9, 2724 (2018).
Simoni, Y. et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018).
Amsen, D., van Gisbergen, K. P. J. M., Hombrink, P. & van Lier, R. A. W. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 19, 538–546 (2018).
Okła K., Farber D. L., Zou W. Tissue-resident memory T cells in tumor immunity and immunotherapy. J. Exp. Med. https://doi.org/10.1084/jem.20201605 (2021).
Luoma, A. M. et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 185, 2918–2935.e29 (2022).
Savas, P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24, 986–993 (2018).
Clarke J. et al. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J. Exp. Med. https://doi.org/10.1084/jem.20190249 (2019).
Corgnac S. et al. CD103+CD8+ TRM cells accumulate in tumors of anti-pd-1-responder lung cancer patients and are tumor-reactive lymphocytes enriched with Tc17. Cell Rep. Med. https://doi.org/10.1016/j.xcrm.2020.100127 (2020).
Masopust, D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010).
Casey, K. A. et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012).
Steinert, E. et al. Quantifying memory CD8+ T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015).
Zhang, N. & Bevan, M. J. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity 39, 687–696 (2013).
Macleod B. L. et al. A network of immune and microbial modifications underlies viral persistence in the gastrointestinal tract. J. Exp. Med. https://doi.org/10.1084/jem.20191473 (2020).
Sandu I. et al. Landscape of exhausted virus-specific CD8 T cells in chronic LCMV infection. Cell Rep. https://doi.org/10.1016/j.celrep.2020.108078 (2020).
Daniel, B. et al. Divergent clonal differentiation trajectories of T cell exhaustion. Nat. Immunol. 23, 1614–1627 (2022).
Ariotti, S. et al. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc. Natl Acad. Sci. USA 109, 19739–19744 (2012).
Schenkel J. M., Fraser K. A., Vezys V., Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. https://doi.org/10.1038/ni.2568 (2013).
Beura, L. K. et al. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19, 173 (2018).
Behr, F. M. et al. Tissue-resident memory CD8+ T cells shape local and systemic secondary T cell responses. Nat. Immunol. 21, 1070–1081 (2020).
Fonseca, R. et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 21, 412–421 (2020).
Swanson, E. et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. eLife 10, e63632 (2021).
Buquicchio, F. A. et al. Distinct epigenomic landscapes underlie tissue-specific memory T cell differentiation. Immunity 57, 2202–2215.e6 (2024).
Wu, J. et al. T cell factor 1 suppresses CD103+ lung tissue-resident memory T cell development. Cell Rep. 31, 107484 (2020).
Fleck, J. S. et al. Inferring and perturbing cell fate regulomes in human brain organoids. Nature 621, 365–372 (2023).
Weber E. W. et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science https://doi.org/10.1126/science.aba1786 (2021).
Schade, A. E. et al. Dasatinib, a small-molecule protein tyrosine kinase inhibitor, inhibits T-cell activation and proliferation. Blood 111, 1366–1377 (2008).
Wherry, E. J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007).
Giles, J. R. et al. Shared and distinct biological circuits in effector, memory and exhausted CD8+ T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 23, 1600–1613 (2022).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
Giles, J. R. et al. Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574.e7 (2022).
Stelekati, E. et al. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity 40, 801–813 (2014).
Ge, C. et al. Bystander activation of pulmonary TRM cells attenuates the severity of bacterial pneumonia by enhancing neutrophil recruitment. Cell Rep. 29, 4236–4244.e3 (2019).
Sheridan, B. S. et al. Oral infection drives a distinct population of intestinal resident memory CD8+ T cells with enhanced protective function. Immunity 40, 747–757 (2014).
Park, S. L., Gebhardt, T. & Mackay, L. K. Tissue-resident memory T cells in cancer immunosurveillance. Trends Immunol. 40, 735–747 (2019).
Edwards, J. et al. CD103+ tumor-resident CD8+ T cells are associated with improved survival in immunotherapy-naive melanoma patients and expand significantly during anti-PD-1 treatment. Clin. Cancer Res. 24, 3036–3045 (2018).
Jaiswal, A. et al. An activation to memory differentiation trajectory of tumor-infiltrating lymphocytes informs metastatic melanoma outcomes. Cancer Cell 40, 524–544.e5 (2022).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Matloubian, M. et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004).
Beura, L. K. et al. T cells in nonlymphoid tissues give rise to lymph-node-resident memory T cells. Immunity 48, 327–338 (2018).
Mackay, L. K. et al. Maintenance of T cell function in the face of chronic antigen stimulation and repeated reactivation for a latent virus infection. J. Immunol. 2012, 2173–2178 (2012).
Zhu, J. et al. Immune surveillance by CD8αα+ skin-resident T cells in human herpes virus infection. Nature 497, 494–497 (2013).
Malik, B. T. et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol. 2, eaam6346 (2017).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Minnie S. A., et al. TIM-3+ CD8 T cells with a terminally exhausted phenotype retain functional capacity in hematological malignancies. Sci. Immunol. https://doi.org/10.1126/sciimmunol.adg1094 (2024).
LaFleur, M. W. et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol. 20, 1335–1347 (2019).
Krishna, S. et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334 (2020).
Guo, Y. et al. Metabolic reprogramming of terminally exhausted CD8(+) T cells by IL-10 enhances anti-tumor immunity. Nat. Immunol. 22, 746–756 (2021).
Lee, Y. J. et al. CD39(+) tissue-resident memory CD8(+) T cells with a clonal overlap across compartments mediate antitumor immunity in breast cancer. Sci. Immunol. 7, eabn8390 (2022).
Virassamy, B. et al. Intratumoral CD8+ T cells with a tissue-resident memory phenotype mediate local immunity and immune checkpoint responses in breast cancer. Cancer Cell 41, 585–601.e8 (2023).
Blackburn, S. D., Shin, H., Freeman, G. J. & Wherry, E. J. Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proc. Natl Acad. Sci. USA 105, 15016–15021 (2008).
Siddiqui, I. et al. Intratumoral Tcf1+ PD-1+ CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211 (2019).
Voabil, P. et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat. Med. 27, 1250–1261 (2021).
Odorizzi, P. M., Pauken, K. E., Paley, M. A., Sharpe, A. & Wherry, E. J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137 (2015).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014).
O’Flanagan, C. H. et al. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 20, 210 (2019).
Abascal, F. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Kartha, V. K. et al. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2, 100166 (2022).
Rauluseviciute, I. et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 52, D174–D182 (2024).
Shen, W.-K. et al. AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 51, D39–D45 (2023).
Nüssing S. et al. Efficient CRISPR/Cas9 gene editing in uncultured naive mouse T cells for in vivo studies. J. Immunol. https://doi.org/10.4049/jimmunol.1901396 (2020).
Acknowledgements
We thank all members of the Wherry Laboratory for helpful discussions and critical analysis of this manuscript. We thank the Penn Cytomics and Cell Sorting Shared Resource Laboratory and Children’s Hospital of Philadelphia Flow Cytometry Core for providing technical support and instrumentation. We thank the Penn Dermatology Skin Biology and Diseases Resource-based Center for providing human skin samples. We thank B. Sheridan (Stony Brook University) for providing the IlnAmut strain of LM-OVA. This work was supported by a Cancer Research Institute Irvington Postdoctoral Fellowship (S.L.P.), National Institutes of Health (NIH) F32 grant (AI181343) (M.M.P.), NIH National Institute of Allergy and Infectious Diseases (NIAID) grant 5F30AI174776 (M.A.S.), University of Pennsylvania Medical Scientist Training Program (M.A.S.), National Science Foundation Graduate Research Fellowship (Y.J.H.), MD fellowship of the Boehringer Ingelheim Fonds (D.B.R.), NIH grant 5T32AR007465-40 (V.F.), University of Pennsylvania Colton Center for Autoimmunity (V.F., C.T.E. and E.J.W.), Dermatology Foundation’s Dermatologist Investigator Research Fellowship (V.F.), NIH T32 grant AR007442 and Parker Institute for Cancer Immunotherapy Scholar Award (J.E.W.), NIH/National Institute of Dental and Craniofacial Research (NIDCR) grant R01DE034056 (D.B.), NIH grants P50CA261608 and R01CA273018 (A.C.H.), NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) K08-AR0802666 (C.T.E.), grant IRG-22-150-41-IRG from the American Cancer Society and the Breakthrough Challenge Foundation (A.D.), NIH/NIAMS grant P30-AR069589 (Penn Skin Biology and Diseases Resource-based Center), NIH grants AI155577; AI115712; AI117950; AI108545; AI082630 and CA210944, the Mark Foundation and Parker Institute for Cancer Immunotherapy (E.J.W.).
Author information
Authors and Affiliations
Contributions
S.L.P. and E.J.W. conceived the study and designed the experiments. S.L.P., M.M.P., V.A., M.M., M.S., D.M., L.T., D.R., N.D., T.C., M.K., Y.J.H., V.F., W.K., S.F.N., A.E.B., J.E.W., M.T., M.A.C. and J.R.G. carried out experiments. C.T.B., E.P., Y.L., K.R., R.M.B, E.R.T., D.B., A.C.H., C.T.E. and A.D. acquired and provided samples for investigation. S.L.P., M.M.P., S.M., R.R.G. and J.R.G. analyzed data. S.L.P. and M.M.P. prepared visualizations. S.L.P. and E.J.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.C.H. received research funding from Bristol Myers Squibb and Merck. C.T.E. holds equity in Cabaletta Bio and has licensed patents with Cabaletta Bio and Novartis. J.R.G. is a consultant for Arsenal Biosciences, Cellanome, Seismic Therapeutics and GVM1. E.J.W. is a member of the Parker Institute for Cancer Immunotherapy which supported this study. E.J.W. is an advisor for Arpelos Bioscience, Arsenal Biosciences, Coherus, Danger Bio, IpiNovyx, New Limit, Marengo, Pluto Immunotherapeutics, Related Sciences, Santa Ana Bio and Synthekine. E.J.W. is a founder of Arpelos Bioscience, Danger Bio and Arsenal Biosciences. E.J.W. holds stock in Coherus. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Tuoqi Wu and the other anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Nick Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotype and function of tissue CD8+ T cells in acute or chronic infection.
a, P14 T cells isolated from tissues 30 dpi with LCMV Arm or Cl13. b, Viral titers 28-32 dpi. c, Marker expression by P14 cells 30 dpi. d, Frequency (†) or absolute number (*) of CD69+CD103+ P14 cells in SG. Data points represent mean; shading represents 95% confidence interval. e, Geometric mean fluorescence intensity (gMFI) of markers in CD69+CD103+ P14 cells 30 dpi from the SI of Arm (TRM) or Cl13 mice, or Arm Spl TCIRCM. f, g, Marker expression by P14 T cells from Spl (f) or SI (g) 30 dpi. h, Frequency of Ly108−CX3CR1− cells within total P14 (Spl), CD69+CXCR6+ P14 (Liv or Kid) or CD69+CD103+ P14 cells (SG, SI) 30 dpi. i, gMFI of markers in CD69+CD103+ P14 cells isolated 30 dpi from the SI of Arm (TRM) or Cl13 mice, or Arm Spl TCIRCM. j, CD39 expression in P14 cells 30 dpi. k, Marker expression in CD69+CD103+ (SI, SG) or CD69+CXCR6+ (Liv) P14 cells from Arm or Cl13-infected mice compared to Arm Spl TCIRCM and Cl13 Spl TEX-TERM 30–40 dpi. Dashed line indicates average in Arm Spl TCIRCM. l, Cytokine production by Arm Spl TCIRCM, CD69+CD103+ P14 from Arm SI (TRM) or Cl13 SI (l, m) or CD69+CD103+ P14 from Arm SG or Cl13 SG (m) or CD69+CXCR6+ Arm Liv or Cl13 Liv 30–40 dpi following in vitro gp33-41 peptide stimulation. n, Ratio of Bim and Bcl2 gMFI in CD69+CD103+ P14 from SI or SG of Arm (TRM) or Cl13 mice 15-25 dpi. Data are pooled from (a, b, d, h, j, m, n) or representative of (c, e, f, g, i, k, l) 2–3 independent experiments with n = 10 (a, c), 7 or 18 (b), 11-18 (d), 4 or 5 (e), 4–5 (f, g), 10 (h), 4 or 5 (i), 10 (j), 4–5 (k), 4-6 (l), 9 or 10 (m) and 6 (n) mice per group.* or † p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 two-tailed Mann–Whitney test (a, c, d, e, h, i, j, m), two-sided Wilcoxon signed-rank test (n). Bars represent the mean.
Extended Data Fig. 2 T cell states across tissues in acute and chronic infection.
a, P14 cells analyzed by TEA-seq in UMAP using RNA expression only (upper panel) or chromatin only (lower panel) colored by sample (Hashtag; HT). b, Distribution of P14 cells in each cluster from each infection (upper panel), tissue (middle panel) or sorted sample (lower panel). c, Weighted nearest neighbor (WNN) UMAP based on combined RNA and ATAC expression in P14 cells colored by ADT (protein, top row), RNA expression (middle row) or gene activity (bottom row). d, Re-clustered WNN UMAP based on combined gene expression and chromatin in P14 cells from SI and Spl samples only, colored by annotated Seurat subcluster (cluster key d-h). e, RNA expression of top 10 marker genes and selected key genes by each SI subcluster and Spl cluster. f,g, Expression of CD103 protein (ADT) and RNA (Itgae), joint RNA expression of Klf2 and S1pr1 or joint ADT CD69 and CXCR6 expression in SI and Spl P14 cells. h, Distribution of cells from SI and Spl assigned to each subcluster. i, Re-clustered WNN UMAP based on combined gene expression and chromatin in P14 cells from SG and Spl samples only, colored by annotated Seurat subcluster (cluster key i-m). j, RNA expression of top 10 marker genes and selected key genes by each SG-derived subcluster and Spl-derived cluster. k, l, Expression of CD103 protein (ADT) and RNA (Itgae), joint RNA expression of Klf2 and S1pr1 or joint ADT CD69 and CXCR6 expression in SI and Spl P14 cells. m, Distribution of cells from SG and Spl assigned to each subcluster. n, Re-clustered WNN UMAP based on combined gene expression and chromatin in P14 cells from Liv and Spl samples only, colored by annotated Seurat subcluster (cluster key n-q). o, RNA expression of top 10 marker genes and selected key genes by each Liv subcluster and Spl-derived cluster. p, Expression of Gzma RNA or joint RNA expression of Klf2 and S1pr1 in Liv and Spl P14 cells. q, Distribution of cells from Liv and Spl assigned to subcluster. Data are pooled from 20-25 mice per infection per tissue. Purple shading in g and l indicates cells annotated as CD103-ADT+.
Extended Data Fig. 3 Transcriptional and epigenetic differences between TRM and TR-TEX cells.
a, Heatmap of RNA expression of genes that are upregulated in Arm TRM versus Arm Spl TCIRCM and are also upregulated in Cl13 TR-TEX versus Cl13 Spl TEX-TERM. b, Violin plots displaying gene expression in indicated Seurat clusters. c, Volcano plot of differentially expressed genes (DEGs) between Arm SI TR-TEX and Cl13 Spl TEX-TERM. d, Volcano plots displaying pairwise DEGs (left column) and rank-ordered plots displaying pairwise differentially accessible chromatin regions (DACRs) (right column) between Arm TRM and Cl13 TR-TEX from SG (top panel) or Liv (bottom panel). e, Number of DEGs (left panel) and DACRs (right panel) between Arm Spl TMEM and Cl13 Spl TEX-TERM or tissue-matched Arm TRM and Cl13 TR-TEX P14 cells. f, Proportion of genes upregulated or downregulated in CD103-ADT+ Arm TRM or CD103-ADT+ Cl13 TR-TEX from the SI or SG that are also up- or downregulated by total Arm TRM or Cl13 TR-TEX from the same tissue. Data are pooled from 20-25 mice per infection per tissue.
Extended Data Fig. 4 Differential regulation of exhaustion, memory and tissue-residency factors in TRM and TR-TEX cells.
a, ATAC coverage plots of DACRs in indicated gene loci for each cluster. SI and SG Arm TRM and Cl13 TR-TEX are subsetted to CD103-ADT+ cells. Green bars indicate DACRs enriched in Arm Spl TMEM compared to Arm TRM, blue bars indicate DACRs enriched in Arm TRM compared to location-matched Cl13 TR-TEX from at least one tissue, pink bars indicate DACRs enriched in Cl13 TR-TEX compared to location-matched Arm TRM from at least one tissue. b, Pairwise transcription factor (TF) motif enrichment in SI (left panel), SG (middle panel) or Liv (right panel) Arm TRM compared to tissue-matched Cl13 TR-TEX (x axis) plotted against pairwise TF motif enrichment in Arm Spl TMEM compared to Cl13 Spl TEX-TERM (y axis). MeanDiff = mean difference determined by chromvar motif deviation analysis. c, RNA expression of indicated TFs (upper panel) and of genes in Pando-defined regulons predicted to be controlled by each TF (positive regulon activity). Data are pooled from 20-25 mice per infection per tissue.
Extended Data Fig. 5 Requirements for Runx3, Blimp1 and Hobit for residency programming across tissues.
a, Ratio of total P14 cells electroporated with indicated sgRNAs versus control Cd19 sgRNAs from Arm or Cl13-infected SI (colored) or spleen (gray) at 8-9 dpi. b, Coexpression of CD69 and CD103 by SI P14 T cells electroporated with control sgCd19 or indicated TF-targeting sgRNAs at 8-9 dpi with Arm or Cl13. c, Ratio of congenically distinct and co-transferred Arm TRM or Cl13 TR-TEX electroporated with identical control Cd19 sgRNAs (same guide in each congenic population) in indicated tissues (SI, SG P14 gated on CD69+CD103+, Liv P14 gated on CD69+CXCR6+) compared to co-transferred total Spl-derived P14 cells at 26-37 dpi. d, Ratio of co-transferred Arm TRM, Cl13 TR-TEX or total Spl-derived P14 cells electroporated with Prdm1 or Runx3 sgRNAs versus control Cd19 sgRNAs at 30-37 dpi. Arm TRM and Cl13 TR-TEX were gated as CD69+CD103+ in the SG and as CD69+CXCR6+ in the Liv. e, Ratio of total CD69+ P14 cells electroporated with indicated sgRNAs versus control Cd19 sgRNAs from Arm or Cl13-infected tissues compared to ratio of total Spl-derived P14 cells at 26-37 dpi. Data are pooled from 2 independent experiments with n = 9 (a, b), 15 (c), 4 or 5 or 9 (d) and 5 or 10 or 13 mice (e) per group.* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 two-sided paired T test (spleen versus tissue P14s) or two-tailed T test (Arm versus Cl13 tissue P14s) (a, c-e).
Extended Data Fig. 6 Tox coordinates TR-TEX but not TRM cell residency programming.
a, Expression of Tox in total sgCd19 and sgTox electroporated P14 cells isolated from the SI at 8-9 dpi with Arm or Cl13. b, Ratio of total P14 cells electroporated with Cd19 control or Tox targeting sgRNAs isolated from the Spl or SI at 8 dpi or >4wks pi with Arm or Cl13. c, Frequency of CD69+CD103+ sgCd19 or sgTox electroporated P14 cells isolated from the SI at 8 dpi. d, Ratio of co-transferred Arm TRM, Cl13 TR-TEX or total Spl-derived P14 cells electroporated with Tox sgRNAs versus control Cd19 sgRNAs >4wks pi. Arm TRM and Cl13 TR-TEX were both gated as CD69+CD103+ in the SG or as CD69+CXCR6+ in the Liv. e, Frequency of P14 cells that were CD69+CD103+ (SI, SG) or CD69+CXCR6+ (Liv) following electroporation with Tox or Cd19 targeting sgRNAs isolated from indicated tissues >4wks pi. f, Ratio of total CD69+ P14 cells electroporated with Tox sgRNAs versus control Cd19 sgRNAs from Arm or Cl13-infected tissues compared to ratio of total P14 cells in the spleen >4wks pi. g, Expression of indicated surface molecules by total P14 cells electroporated with indicated sgRNAs and isolated from the SI 8-9 dpi with Arm or Cl13. h, Fold change (FC) in expression of indicated surface molecules in total P14 cells electroporated with sgRNAs directed towards genes encoding indicated TFs compared to control cells electroporated with sgRNAs directed towards Cd19 at 8-9 dpi with Arm or Cl13. Data are pooled from 2–3 experiments with n = 24 (b, c), 22 (d, e, f) or 8-24 mice (g, h). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 two-tailed paired T test (spleen versus tissue P14s) or two-tailed T test (Arm versus Cl13 tissue P14s) (b-f) or two-sided Mann–Whitney test (h). Bars represent the mean.
Extended Data Fig. 7 Developmental plasticity and lineage relationships between TRM and TEX cells.
a, CD107a in donor P14 cells rechallenged with Arm or Cl13 (or non-rechallenged TRM) 21–28 dpi following ex vivo gp33-41 stimulation. Statistics indicate tests between same donor origin. b, Total P14 cells in Spl after Cl13 rechallenge. c, Phenograph (pg) clustering of donor P14 cells from rechallenged mice. d, Marker expression in pg clusters; TEX-TERM (purple). e, Proportion of Spl TEX-TERM in pg clusters after Cl13 rechallenge. f, g, Marker expression by Spl TEX-TERM after Cl13 compared to Arm rechallenge. h, Number of total P14 (Spl) or CD69+CD103+ P14 cells (SG) derived from donor populations after Arm rechallenge. i, Proportion of Arm-rechallenged P14 cells CD69+CD103+ in SG. j,k Marker expression by Arm-rechallenged P14 cells in Spl or SI. l, m, Proportion of Arm-rechallenged donor P14 cells CD8a-BV650+ i.v. Bl; blood. n, Number of P14 cells (Spl) or CD69+CD103+ P14 cells (SG) following Cl13 rechallenge. o–r Marker expression by Cl13-rechallenged donor P14 cells. s, Fold change coexpression CD69 and CD103 by rechallenged P14 cells compared to naive-derived SI P14. t, Experiment schematic. u, Number total P14 cells in Spl. v, Number of CD69+CD103+ P14 cells in SI or SG. w, Frequency of P14 cells coexpressing CD69 and CD103 in SI and SG. Dashed lines indicate threshold for inclusion (h, n) or average expression in naive P14-derived cells (Spl) (l, q). Data pooled from or representative of 2 (a, h-r) or 3 (b-g, s-w) independent experiments, with n = 3 or 4 (a), 12 or 15 (b), 9 or 24 (c-e), 7 or 8 or 9 or 10 (f, g), 10 (h-m), 4 or 7 or 8 or 10 or 11 or 12 or 13 or 14 or 16 (n), 7 or 11 or 16 (o), 7, 11 or 17 (p, q), 7 or 10 or 11 or 17 (r, s) and 15 or 20 or 21 (u-w) mice per group. TN; naive P14 cells. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 two-tailed Mann–Whitney Test (a-s, w) or two-sided unpaired parametric T test (u, v). Bars = mean, error bars = SD.
Extended Data Fig. 8 Cell-state specification of TRM and TR-TEX cells.
a, UMAP embedding of inferred single-cell Gene Regulatory Network (GRN) based on combined transcription factor (TF) expression and motif accessibility in all non-naive P14 cells analyzed by TEA-sequencing with expanded gene labeling. Size of nodes (genes) represents the number of connections in network. b, Individual UMAP embeddings of genes and gene modules from inferred single-cell Gene Regulatory Networks (GRNs) engaged in each T cell subset. Node size and color scale represent degree of RNA expression for each gene in network. c, d Enrichment for GRN modules in merged TRM and TR-TEX Seurat clusters from all tissues, or in Arm Spl TMEM and Cl13 Spl TEX-TERM clusters. FC = fold change, δ = Cliff’s delta effect size. Circle size and heat scale indicate relative enrichment per cluster. Boxes in c show the interquartile range (25th–75th percentile), center line indicates the median, and whiskers extend to 1.5x the IQR. e, Heatmap of RNA expression of genes commonly upregulated in Arm TRM and Cl13 TR-TEX versus Arm Spl TCIRCM (pooled Arm Spl TMEM and Arm Spl TEFF clusters). f, Heatmap of RNA expression of genes uniquely upregulated in Arm Spl TCIRCM versus Cl13 Spl TEX-TERM but not in Arm TRM versus Cl13 TR-TEX (left panel), or commonly upregulated in Arm Spl TCIRCM versus Cl13 Spl TEX-TERM and Arm TRM versus Cl13 TR-TEX (right panel). g, Comparative analysis of RNA expression of genes upregulated in 1) Cl13 Spl TEX-TERM versus Arm Spl TCIRCM, 2) Cl13 Spl TR-TEX versus Arm TRM from each tissue, and 3) TRM from each tissue versus Arm Spl TCIRCM. Heatmaps display genes that are uniquely enriched in TEX (top panel) or are also unregulated by Arm TRM (bottom heatmap) compared to Spl TCIRCM. Data are pooled from 20-25 mice per infection per tissue. **** p < 0.001 two-sided unpaired Wilcox test.
Extended Data Fig. 9 Defining cell state-specific gene and surface protein signatures of TRM and TR-TEX cells.
a, Relative RNA expression of genes comprising TRM cell-specific or TR-TEX cell-specific signatures. b, Seurat enrichment for Mackay 2016 TRM core4 and Giles 202263 TEX-TERM signatures in TRM and TR-TEX compared to newly derived TRM and TR-TEX cell-state-specific signatures in LCMV TEA-seq clusters. c, Cliff’s delta effect size and fold change of Seurat enrichment for Mackay 2016 TRM core4 and Giles 202263 TEX-TERM signatures and newly derived TRM and TR-TEX cell-state-specific signatures in LCMV TEA-seq clusters. d, Seurat enrichment for cell-state-specific TRM or TR-TEX signature scores within CD8+ T cells isolated from human SI epithelium or non-naive CD8+ T cells from healthy donor blood65 or GSEA enrichment for cell-state-specific signatures within CD39+PD-1+ TEX cells compared to all non-naive CD8+ T cells from healthy donor blood66. δ = Cliff’s delta, FC = fold change. Boxes = interquartile range (25th–75th percentile), center line = median, whiskers = 1.5x the IQR. e, RNA expression of genes encoding indicated surface proteins in LCMV TEA-seq clusters. Size = proportion of cells expressing each gene; scale bar = relative expression. f, Marker expression in Arm Spl TCIRCM, Cl13 Spl TEX-PROG or Cl13 Spl TEX-TERM cells or TRM and TR-TEX cells from SI (CD69+CD103+) or Liv (CD69+CXCR6+) 30–40 dpi with Arm or Cl13. g, Proportion of TRM or TR-TEX cells expressing CD73 and CD200R in Liv (CD69+CXCR6+) or SG (CD69+CD103+) after Arm or Cl13 infection (left and middle panel) or proportion of TRM-like cells in human epidermal or melanoma samples (KLRG1−CD69+CD103+) expressing CD73 and CD200R (right panel). h, IR expression by total P14 Arm TCIRCM (Spl) or by CD69+CD103+CD73+ TRM cells or CD69+CD103+CD73−CD200R+ TR-TEX cells 30–40 dpi with Arm or Cl13. i, IR expression by KLRG1−CD69+CD103+CD73+ TRM-like cells from human tonsil or epidermis or KLRG1−CD69+CD103+CD73−CD200R+ TRM-like TIL from HNSCC or melanoma. Data pooled from 3–6 human donors (g, i) or representative of at least 2 experiments with 4 or 5 mice per group (e, f, g). TEA-seq data pooled from 20-25 mice per infection per tissue. * p < 0.05, two-tailed Mann–Whitney test. Bars represent the mean.
Extended Data Fig. 10 TRM and TR-TEX cells differentially contribute to immune responses.
a, Frequency of CD69+CD103+ LM-OVA-generated OT-I cells in SI after LCMV or no LCMV infection (no inf). b, Serum viral titers in LM-OVA immune mice 30 dpi with LCMV. LOD = limit of detection. c, Proportion CD69+CD103+ OT-I TRM (LM-derived), P14 TRM (Arm-derived) or P14 TR-TEX (Cl13-derived) expressing CD73 or CD200R after LCMV or no LCMV (LM only). d, e Overall patient survival from TCGA metastatic melanoma (d) or TNBC METABRIC (e) stratifying by TRM and TR-TEX specific signatures (hi = top 25%, lo = bottom 25%) within all patients or the top 50% of CD8hi patients. f, Correlation analysis comparing TRM and TR-TEX signature scores and CD8A expression in TCGA melanoma patients. g, PD-1 expression in CD69+CXCR6+ Arm Kid P14 TRM, CD69+CXCR6+ Cl13 Kid P14 TR-TEX or total Spl Arm P14 TCIRCM. Dotted line = Arm P14 TCIRCM mean. h, PD-1 gMFI in CD69+CD103+ Arm SI P14 TRM or Cl13 TR-TEX and PD-L1 in MHCII+CD11c+ SI-derived dendritic cells (DCs) at baseline and 48 h after gp33-41 peptide i.v. i, Number of P14 cells in SI after FTY720 and α-PD-L1 treatment and/or i.v. gp3333-41. j, k, Cytokine production by ex vivo gp3333-41 restimulated total Spl P14 cells (i) or CD69+CXCR6+ Cl13 Spl TEX-TERM (j) treated with α-PD-L1 and/or i.v. gp3333-41. l-o Cytokine production or degranulation by ex vivo gp3333-41 restimulated CD69+CD103+ SI P14 TRM (Arm) or TR-TEX (Cl13) after α-PD-L1 treatment with gp33-41 i.v. Data pooled from or representative of 3 independent experiments (a-c) or 2–3 independent experiments (g-o) with n = 8 or 10, 4 or 5 or 6 (g, h), 4 or 5 or 8 or 9 or 11 (i), 9 or 11 or 12 or 13 (j), 12 (k), 11-12 (l), 3 or 4 or 5 or 6 or 7 (m) 12 or 13 or 14 or 15 (n, o) mice per group. * p < 0.05, ** p < 0.01, *** p < 0.001, two-tailed Kruskal–Wallis test (a-c, h), two-tailed Mann–Whitney test (i-o), Kaplan–Meier estimate (d, e) or Pearson correlation test (f). Shading (d, e) = 50% confidence interval, solid line = Kaplan–Meier estimate. Bars = mean.
Supplementary information
Supplementary Information
Flow cytometry gating strategies used in the study.
Supplementary Table 1
Shared and distinct genes up- or downregulated in TRM and TR-TEX compared to TCIRCM cells.
Supplementary Table 2
Pairwise DEGs between TRM and TR-TEX in each tissue.
Supplementary Table 3
Pairwise DACRs between TRM and TR-TEX in each tissue.
Supplementary Table 4
TRM and TR-TEX cell-state-specific gene signatures.
Supplementary Table 5
Human participant information.
Supplementary Table 6
List of antibodies used in the study.
Supplementary Table 7
Custom oligos and sgRNAs used in the study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, S.L., Painter, M.M., Manne, S. et al. Tissue-resident exhausted and memory CD8+ T cells have distinct ontogeny, function and roles in disease. Nat Immunol 27, 110–125 (2026). https://doi.org/10.1038/s41590-025-02352-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02352-y