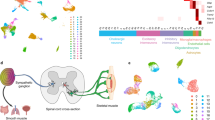
Abstract
Neurogenesis lasts ~10 times longer in developing humans compared to mice, resulting in a >1,000-fold increase in the number of neurons in the CNS. To identify molecular and cellular mechanisms contributing to this difference, we studied human and mouse motor neurogenesis using a stem cell differentiation system that recapitulates species-specific scales of development. Comparison of human and mouse single-cell gene expression data identified human-specific progenitors characterized by coexpression of NKX2-2 and OLIG2 that give rise to spinal motor neurons. Unlike classical OLIG2+ motor neuron progenitors that give rise to two motor neurons each, OLIG2+/NKX2-2+ ventral motor neuron progenitors remain cycling longer, yielding ~5 times more motor neurons that are biased toward later-born, FOXP1-expressing subtypes. Knockout of NKX2-2 converts ventral motor neuron progenitors into classical motor neuron progenitors. Such new progenitors may contribute to the increased production of human motor neurons required for the generation of larger, more complex nervous systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
scRNA-seq datasets produced in this study are available in the GEO under GSE270069. Cell lines (ES and iPS), along with plasmids and viral constructs, will be made freely available to investigators at academic institutions for noncommercial research upon request. Public datasets for mouse in vivo single-cell RNA-seq data are available under accession E-MTAB-7320. Human in vivo single-cell RNA-seq data are available in the GEO under accessions GSE171890 and GSE219122. The TcoF database is accessible via https://tools.sschmeier.com/tcof/home/.
Code availability
Code used in this study is available at https://github.com/wichterle-lab/.
References
Braitenberg, V. & Braitenberg, V. Brain size and number of neurons: an exercise in synthetic neuroanatomy. J. Comput. Neurosci. 10, 71–77 (2001).
Stepien, B. K., Vaid, S. & Huttner, W. B. Length of the neurogenic period-a key determinant for the generation of upper-layer neurons during neocortex development and evolution. Front. Cell Dev. Biol. 9, 676911 (2021).
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009).
Hill, R. S. & Walsh, C. A. Molecular insights into human brain evolution. Nature 437, 64–67 (2005).
Miller, D. J., Bhaduri, A., Sestan, N. & Kriegstein, A. Shared and derived features of cellular diversity in the human cerebral cortex. Curr. Opin. Neurobiol. 56, 117–124 (2019).
Götz, M. & Huttner, W. B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005).
Franchini, L. F. Genetic mechanisms underlying cortical evolution in mammals. Front. Cell Dev. Biol. 9, 591017 (2021).
Florio, M., Borrell, V. & Huttner, W. B. Human-specific genomic signatures of neocortical expansion. Curr. Opin. Neurobiol. 42, 33–44 (2017).
Hansen, D. V., Lui, J. H., Parker, P. R. L. & Kriegstein, A. R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010).
Johnson, M. B. et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646 (2015).
Pollen, A. A. et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015).
Kanning, K. C., Kaplan, A. & Henderson, C. E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440 (2010).
Stifani, N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci. 8, 293 (2014).
Rayon, T. et al. Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667 (2020).
MacLarnon, A. The evolution of the spinal cord in primates: evidence from the foramen magnum and the vertebral canal. J. Hum. Evol. 30, 121–138 (1996).
Meyer, M. R. The spinal cord in hominin evolution. eLS https://doi.org/10.1002/9780470015902.a0027058 (2016).
Nornes, H. O. & Carry, M. R. Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 159, 1–6 (1978).
Wichterle, H., Lieberam, I., Porter, J. A. & Jessell, T. M. Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 (2002).
Maury, Y. et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 (2014).
Marklund, U. et al. Detailed expression analysis of regulatory genes in the early developing human neural tube. Stem Cells Dev. 23, 5–15 (2014).
Sansom, S. N. et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 5, e1000511 (2009).
Sun, T. et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr. Biol. 11, 1413–1420 (2001).
Novitch, B. G., Chen, A. I. & Jessell, T. M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789 (2001).
Arber, S. et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23, 659–674 (1999).
Tsuchida, T. et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79, 957–970 (1994).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Zhang, X. et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100 (2010).
Rayon, T., Maizels, R. J., Barrington, C. & Briscoe, J. Single-cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human-specific features. Development 148, dev199711 (2021).
Delile, J. et al. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146, dev173807 (2019).
Li, X. et al. Profiling spatiotemporal gene expression of the developing human spinal cord and implications for ependymoma origin. Nat. Neurosci. 26, 891–901 (2023).
Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022).
Kawaguchi, A. et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135, 3113–3124 (2008).
Kageyama, R., Ohtsuka, T. & Kobayashi, T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 (2007).
Amoroso, M. W. et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574–586 (2013).
Thiry, L., Hamel, R., Pluchino, S., Durcan, T. & Stifani, S. Characterization of human iPSC-derived spinal motor neurons by single-cell RNA sequencing. Neuroscience 450, 57–70 (2020).
Hollyday, M. & Hamburger, V. An autoradiographic study of the formation of the lateral motor column in the chick embryo. Brain Res. 132, 197–208 (1977).
Prasad, A. & Hollyday, M. Development and migration of avian sympathetic preganglionic neurons. J. Comp. Neurol. 307, 237–258 (1991).
Whitelaw, V. & Hollyday, M. Thigh and calf discrimination in the motor innervation of the chick hindlimb following deletions of limb segments. J. Neurosci. 3, 1199–1215 (1983).
Briscoe, J. et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398, 622–627 (1999).
Iwata, R. & Vanderhaeghen, P. Regulatory roles of mitochondria and metabolism in neurogenesis. Curr. Opin. Neurobiol. 69, 231–240 (2021).
Diaz-Cuadros, M. et al. In vitro characterization of the human segmentation clock. Nature 580, 113–118 (2020).
Matsuda, M. et al. Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129 (2020).
Workman, A. D., Charvet, C. J., Clancy, B., Darlington, R. B. & Finlay, B. L. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci. 33, 7368–7383 (2013).
Borrell, V. & Götz, M. Role of radial glial cells in cerebral cortex folding. Curr. Opin. Neurobiol. 27, 39–46 (2014).
Fiddes, I. T. et al. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018).
Suzuki, I. K. et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16 (2018).
Raballo, R. et al. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012–5023 (2000).
Wang, L., Hou, S. & Han, Y.-G. Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat. Neurosci. 19, 888–896 (2016).
Chenn, A. Wnt/beta-catenin signaling in cerebral cortical development. Organogenesis 4, 76–80 (2008).
Ying, Q.-L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Schmeier, S., Alam, T., Essack, M. & Bajic, V. B. TcoF-DB v2: update of the database of human and mouse transcription co-factors and transcription factor interactions. Nucleic Acids Res. 45, D145–D150 (2017).
Miao, Z., Deng, K., Wang, X. & Zhang, X. DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics 34, 3223–3224 (2018).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Acknowledgements
We thank G. Yeo and D. Gifford for help with and access to their shared computing server, E. Bush and M. Finlayson of the Columbia University Sulzberger Genome Center for running the 10x single-cell library preparation and sequencing, and M. Kissner and the CSCI Stem Cell Facility for assistance with flow cytometry. We thank C. Marchetto for kindly sharing Rhesus macaque iPS cells and culture conditions. We thank S. DiIorio for designing and testing sequencing primers for barcode analysis. We are grateful to C. Mason, F. Polleux and M. Closser and members of the Wichterle, Chaolin Zhang and Edmund Au labs for discussion and feedback. H.W. holds an endowed chair from Jerry and Emily Spiegel. We acknowledge grants from the NIH (R01NS116141 and R01NS089676 to H.W.; K99MH130892 to S.J.) and Project ALS (to H.W.) and a seed grant from the Columbia Stem Cell Initiative (to S.J.).
Author information
Authors and Affiliations
Contributions
Conceptualization: S.J. and H.W. Experiments: S.J. Data analysis: S.J. Human NKX2-2-CreERT2 iPS cell line generation: E.G. Writing—original draft: S.J. and H.W. Writing—review and editing: S.J. and H.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Shen-Ju Chou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of human-specific progenitors in motor neurogenesis.
(a) Culture conditions for motor neuron differentiation. (b) UMAP of all high-quality single-cell RNA-seq profiles after removal of interneuron-lineage/fibroblast-like cells, colored based on replicate. (c) Timestamp distribution in human single-cell gene expression datasets across the two replicates, normalized by cell cluster (day 10 cells in replicate 1 were not timestamped and therefore inferred based on absence of timestamp expression; see Methods) (d) Timestamp distribution (day 4–6) in mouse single-cell gene expression datasets across the two replicates. (Day 7 was not collected for first replicate.) (e) UMAP of all high-quality single-cell RNA-seq profiles, clusters colored based on identity. (f) UMAP of combined human and mouse single-cell gene expression profiles following CCA-mediated integration, colored by normalized expression level of key marker genes. (g) Chi-square distance between human and mouse clusters. Smaller distances indicate greater similarity in constituent cells’ distribution across common clusters. (h) Classification of human (top) and mouse (bottom) cells according to human-based random-forest classifier shows that the vast majority of mouse cells classified as H0-2 (pMN), H6 (late-appearing NKX2-2+/OLIG2+), or H7-9 (MN), leaving the H3-5 (vpMN) category void of classified mouse cells. (i) Macaque iPSC cultures display NKX2-2 and OLIG2 co-expressing cells when ISL1+ MNs begin to appear, mimicking human cultures (scale bar = 50μm). (j) UMAP of scRNA-seq data from Carnegie Stage 12 human embryonic spinal cords30, colored based on cluster identity or NKX2-2/OLIG2 expression. (k) Left: Alignment of human in vitro and human CS12 embryonic scRNA-seq data shows that human-specific clusters map onto distinct clusters found in vivo. Right: Alignment of human in vitro and mouse E9.5-10.5 spinal cord shows that H4 cells show poor overlap with all embryonic mouse clusters, suggesting that H4-like cells are found in human (but not mouse) embryonic spinal cords. (l) Normalized expression levels of key marker genes in CS12 human (top) and E9.5-E10.5 mouse (bottom) clusters. (m) Alignment of human in vitro and human CS14/pcw5 (CS14) embryonic scRNA-seq data. (n) Normalized expression levels of key marker genes in CS14/pcw5 human clusters.
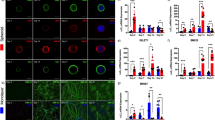
Extended Data Fig. 2 Characterization of NKX2-2-dependent lineage tracing.
(a) Left: varying lengths and concentrations of 4OHT pulse affect the efficiency of recombination-based RFP expression (mean ± SD; two-way ANOVA; n = 3 biological replicates; time \(p < 1\times {10}^{-4}\); concentration \(p=0.03\)). Right: proportions of NKX2-2-positive cells within RFP-positive populations on day 13, 48 hours post 4OHT removal (two-way ANOVA; n = 3 biological replicates). (b) The same panel of cells as shown in Fig. 2c, immunolabeled for OLIG2 and NKX2-2 and counter-stained with DAPI (scale bar = 50μm). (c) Day 12 human embryoid bodies differentiated with and without SAG (Smoothened agonist), and with 4OHT pulse between days 9-11 show that in the absence of SAG, NKX2-2 expression as well as Cre-dependent recombination is lost (scale bar = 50μm). (d) Representative flow cytometry gating strategy to remove debris and clumps of cells. (e) Day 16 human culture immunostained for pan-neuronal marker NEUN and ISL1 shows that the vast majority of neurons produced are motor neurons (scale bar = 50μm). (f) Flow cytometry analysis of day 16 human cultures shows that the vast majority of ISL1-positive cells are MNX1-positive and vice versa in both RFP+ and RFP− populations (representative differentiation, n = 1, sample pooled across 50+ EBs). (g) Human cultures following 4OHT treatment on days 9–11 show that many RFP+ cells retain progenitor identity (OLIG2+ or OLIG2+/NKX2-2+) on day 14; however, with DAPT treatment, virtually no cells express OLIG2 or NKX2-2 (scale bar = 50μm). (h) Majority of cells in human cultures express ISL1 in response to DAPT, with a small fraction (<1%) of cells expressing V2 interneuron marker CHX10 (scale bar = 50μm). (i) UMAP of all high-quality human scRNA-seq profiles, colored based on expression of V2 and V3 interneuron markers (CHX10 and SIM1), fibroblast marker (COL1A2), and timestamp identity.
Extended Data Fig. 3 vpMNs display delayed and protracted neurogenesis.
(a) Top enriched pathways in Reactome analysis of genes upregulated in vpMN relative to pMN (q-value\(\,\le\) 0.001). (b) Differential gene expression analysis between in vivo vpMN (HE4) and pMN (HE1) clusters (Supplemental figure 1J) shows that, similar to in vitro, vpMNs display characteristics of higher Notch activity relative to pMNs (DEsingle). (c) Schematic of cumulative BrdU labeling assay to derive MN birthcurve for vpMN and pMN lineages. (d) Changes in the proportion of BrdU+, day 21 MNs for vpMN (RFP+), total and pMNIMPUTED lineages following cumulative BrdU labeling at progressively later timepoints (mean ± SD; n = 3 biological replicates). Despite all three curves starting at close to 100% (indicating that most vpMNs and pMNs are mitotic at day 9), the vpMN curve is right-shifted, indicating that RFP+ vpMNs remain mitotic for longer. (e) Flow cytometry plots for day 21 human cultures (only ISL1/2+ MNs shown) treated with BrdU starting at progressively later timepoints show slower decrease in BrdU+ MNs in RFP+ compared to RFP− populations. (f) Estimation of cell cycle length based on EdU pulse-labeling shows that RFP+ have similar cell cycle length compared to the total cells (mean ± SD; unpaired two-sided t-test; n = 3 biological replicates; \(p=0.07\)). (g) RFP+ and RFP- cells display similar proportions of cleaved Caspase-3−positive progenitors and motor neurons, indicating that cell death rates are similarly low for vpMN and pMN lineages (mean ± SD; unpaired two-sided t-test; n = 3 biological replicates; from left to right: \(p=0.36,\,0.29\)). (h) Proportion of RFP+ cells within newborn motor neuron population (as determined by BrdU labeling) increases over time, indicating that vpMNs undergo delayed neurogenesis (mean ± SD; n = 3 biological replicates). (i) Numbers of motor neuron progenitors and motor neurons at the onset of and tail-end of neurogenesis in both mouse and human, numbers for each replicate shown separately. (j) Day 18 human cultures immunostained for FOXP1, LHX3 and MNX1 show that FOXP1+ cells are LHX3− and MNX1+, indicating that they are LMC-like MNs (scale bar = 50 µm). (k) Day 18 human cultures immunostained for FOXP1, LHX1 and ISL1 show that LHX1 is expressed in a subset of FOXP1 cells but is not co-expressed with ISL1, suggesting that FOXP1+/LHX1+ cells are LMC-l-like MNs (scale bar = 50μm). (l) Human cultures show sequential appearance of MMC/HMC-, LMCm- and LMCl-like MNs (mean ± SD; chi-squared test; n = 3 biological replicates; \(p < 1\times {10}^{-5}\)). (m) Cumulative BrdU labeling, followed by immunolabeling for FOXP1 and ISL1/2 reveal that the birthcurve of FOXP1+ MNs is slightly late-shifted relative to the total birthcurve (mean ± SD; unpaired two-sided t-test; n = 3 biological replicates; ** from left to right: \(p=0.003,\,0.001\)). (n) Early (day 8) treatment with DAPT produces ~99% FOXP1− MNs (left), supporting the finding that early-born MNs are void of FOXP1-expressing subtypes. In contrast, adding DAPT on day 10 results in ~30% FOXP1+ MNs (right).
Extended Data Fig. 4 NKX2-2 knockout functionally reverts vpMNs to pMNs.
(a) Both WT and NKX2-2 KO cultures display broad OLIG2 expression and begin to show ISL1+ MNs at day 10 (scale bar = 50μm). (b) NKX2-2 KO cells lack NKX2-2 immuno-reactivity in both RFP+ and RFP− cells (scale bar = 50μm). (c) GFP-expressing WT cells can easily be distinguished from NKX2-2 KO cells in mosaic co-cultures based on flow cytometry. (d) Schematic for BrdU pulse-labeling. (e) Proportion of MNs born after day 17 (assessed at day 19) in WT and NKX2-2 KO MNs (RFP+ and total) show an even greater fold-change between WT and KO cells (~2.5-fold, mean ± SD; unpaired two-sided t-test; n = 3 biological replicates; *** \(p < 1\times {10}^{-4}\); n.s. \(p=0.27\); ** \(p < 0.01\)). (f) The proportion of GFP- NKX2-2 KO cells decreases over time in co-cultures, both in total and MN populations (mean ± SD; n = 3 biological replicates). (g) Relative numbers of progenitors and MNs in WT and NKX2-2 KO populations in mosaic co-cultures at the beginning (day 11) and end (day 19) of neurogenesis, normalized to number of WT progenitors on day 11 (mean ± SD; n = 5 biological replicates).
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jang, S., Gumnit, E. & Wichterle, H. A human-specific progenitor sub-domain extends neurogenesis and increases motor neuron production. Nat Neurosci 27, 1945–1953 (2024). https://doi.org/10.1038/s41593-024-01739-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-024-01739-8