Abstract
Brain function relies on the generation of a large variety of morphologically and functionally diverse, but specific, neuronal synapses. Here we show that, in mice, the initial formation of synapses on cerebellar Purkinje cells involves a presynaptic protein—CBLN1, a member of the C1q protein family—that is secreted by all types of excitatory inputs. The molecular program then evolves only in one of the Purkinje cell inputs, the inferior olivary neurons, with the additional expression of the presynaptic secreted proteins C1QL1, CRTAC1 and LGI2. These molecules work in concert to specify the mature connectivity pattern on the Purkinje cell target. These results show that some inputs actively and gradually specify their synaptic molecular identity, while others rely on the ‘original molecular code’. Thus, the molecular specification of excitatory synapses, crucial for proper circuit function, is acquired in a stepwise manner during mouse postnatal development and obeys input-specific rules.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the main text or the Supplementary Information. Single-cell expression data in cerebellar GCs are available at https://apps.kaessmannlab.org/sc-cerebellum-transcriptome/. AlphaFold structure predictions are available on the database (https://alphafold.ebi.ac.uk/). Information on the Kir2.1 channel is available at https://channelpedia.epfl.ch/wikipages/42/. Source data are provided with this paper—statistical source data for figures and extended data figures, and raw image for Extended Data Fig. 5.
Code availability
Plugins are available on GitHub via the following links: custom-made plugin for unbiased 3D detection of individual RNA puncta and DAPI nucleus (https://github.com/orion-cirb/RNA_Scope); custom-made plugin developed based on the 3D Weka Segmentation plugin to allow semi-automatic detection of GFP+ CFs and to count and measure VGLUT2 clusters (https://github.com/orion-cirb/Vglut2_GFP_Weka_Maela.git). Gene clustering was performed using the open-source software Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).
References
Sanes, J. R. & Zipursky, S. L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556 (2020).
Cizeron, M. et al. A brainwide atlas of synapses across the mouse life span. Science 369, 270–275 (2020).
Zhu, F. et al. Architecture of the mouse brain synaptome. Neuron 99, 781–799 (2018).
Sanes, J. R. & Yamagata, M. Many paths to synaptic specificity. Annu. Rev. Cell Dev. Biol. 25, 161–195 (2009).
De Wit, J. & Ghosh, A. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 22–35 (2016).
Hassan, B. A. & Hiesinger, P. R. Beyond molecular codes: simple rules to wire complex brains. Cell 163, 285–291 (2015).
Südhof, T. C. Towards an understanding of synapse formation. Neuron 100, 276–293 (2018).
Changeux, J.-P. & Danchin, A. Selective stabilisation of developing synapses as a mechanism for the specification of neuronal networks. Nature 264, 705–712 (1976).
Sperry, R. W. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl Acad. Sci. USA 50, 703–710 (1963).
Schroeder, A. et al. A modular organization of LRR protein-mediated synaptic adhesion defines synapse identity. Neuron 99, 329–344 (2018).
Sigoillot, S. M. et al. The secreted protein C1QL1 and its receptor BAI3 control the synaptic connectivity of excitatory inputs converging on cerebellar Purkinje cells. Cell Rep. 10, 820–832 (2015).
Uemura, T. et al. Trans-synaptic interaction of GluRδ2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 (2010).
Früh, S. et al. Neuronal dystroglycan is necessary for formation and maintenance of functional CCK-positive basket cell terminals on pyramidal cells. J. Neurosci. 36, 10296–10313 (2016).
Fossati, M. et al. Trans-synaptic signaling through the glutamate receptor δ-1 mediates inhibitory synapse formation in cortical pyramidal neurons. Neuron 104, 1081–1094 (2019).
Heller, E. A. et al. The biochemical anatomy of cortical inhibitory synapses. PLoS ONE 7, e39572 (2012).
Selimi, F., Cristea, I. M., Heller, E., Chait, B. T. & Heintz, N. Proteomic studies of a single CNS synapse type: the parallel fiber/Purkinje cell synapse. PLoS Biol. 7, e1000083 (2009).
Matsuda, K. et al. Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science 328, 363–368 (2010).
Favuzzi, E. et al. Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science 363, 413–417 (2019).
Ito, M. Control of mental activities by internal models in the cerebellum. Nat. Rev. Neurosci. 9, 304–313 (2008).
Buckner, R. L. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80, 807–815 (2013).
Konnerth, A., Llano, I. & Armstrong, C. M. Synaptic currents in cerebellar Purkinje cells. Proc. Natl Acad. Sci. USA 87, 2662–2665 (1990).
Ito, M. & Kano, M. Long-lasting depression of parallel fiber-Purkinje cell transmission induced by conjunctive stimulation of parallel fibers and climbing fibers in the cerebellar cortex. Neurosci. Lett. 33, 253–258 (1982).
Ichikawa, R. et al. Territories of heterologous inputs onto Purkinje cell dendrites are segregated by mGluR1-dependent parallel fiber synapse elimination. Proc. Natl Acad. Sci. USA 113, 2282–2287 (2016).
Hirai, H. et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci. 8, 1534–1541 (2005).
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299 (2016).
Kakegawa, W. et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron 85, 316–329 (2015).
Ito-Ishida, A. et al. Cbln1 regulates rapid formation and maintenance of excitatory synapses in mature cerebellar Purkinje cells in vitro and in vivo. J. Neurosci. 28, 5920–5930 (2008).
Veleanu, M. et al. Modified climbing fiber/Purkinje cell synaptic connectivity in the cerebellum of the neonatal phencyclidine model of schizophrenia. Proc. Natl Acad. Sci. USA 119, e2122544119 (2022).
Sotelo, C. Cellular and genetic regulation of the development of the cerebellar system. Prog. Neurobiol. 72, 295–339 (2004).
Kano, M., Watanabe, T., Uesaka, N. & Watanabe, M. Multiple phases of climbing fiber synapse elimination in the developing cerebellum. Cerebellum 17, 722–734 (2018).
Kita, Y., Tanaka, K. & Murakami, F. Specific labeling of climbing fibers shows early synaptic interactions with immature Purkinje cells in the prenatal cerebellum. Dev. Neurobiol. 75, 927–934 (2015).
Mason, C. A., Christakos, S. & Catalano, S. M. Early climbing fiber interactions with Purkinje cells in the postnatal mouse cerebellum. J. Comp. Neurol. 297, 77–90 (1990).
Altman, J. Postnatal development of the cerebellar cortex in the rat. I. The external germinal layer and the transitional molecular layer. J. Comp. Neurol. 145, 353–397 (1972).
Stoeckli, E. T. Neural circuit formation in the cerebellum is controlled by cell adhesion molecules of the contactin family. Cell Adh. Migr. 4, 523–526 (2010).
Mätlik, K., Govek, E.-E., Paul, M. R., Allis, C. D. & Hatten, M. E. Histone bivalency regulates the timing of cerebellar granule cell development. Genes Dev. 37, 570–589 (2023).
Zhu, X. et al. Role of Tet1/3 genes and chromatin remodeling genes in cerebellar circuit formation. Neuron 89, 100–112 (2016).
Edmondson, J., Liem, R., Kuster, J. & Hatten, M. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J. Cell Biol. 106, 505–517 (1988).
Sepp, M. et al. Cellular development and evolution of the mammalian cerebellum. Nature 625, 788–796 (2024).
Luo, Y. et al. Neurogenic timing of the inferior olive subdivisions is related to the olivocerebellar projection topography. Sci. Rep. 13, 7114 (2023).
Apóstolo, N. et al. Synapse type-specific proteomic dissection identifies IgSF8 as a hippocampal CA3 microcircuit organizer. Nat. Commun. 11, 5171 (2020).
Noya, S. B. et al. The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science 366, eaav2642 (2019).
Fremeau, R. T. et al. The expression of vesicular glutamate transporters defines two classes of excitatory synapse. Neuron 31, 247–260 (2001).
Hioki, H. et al. Differential distribution of vesicular glutamate transporters in the rat cerebellar cortex. Neuroscience 117, 1–6 (2003).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Wang, X., Zhang, C., Szábo, G. & Sun, Q.-Q. Distribution of CaMKIIα expression in the brain in vivo, studied by CaMKIIα-GFP mice. Brain Res. 1518, 9–25 (2013).
Takano, T. et al. Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature 588, 296–302 (2020).
Chen, L. Y., Jiang, M., Zhang, B., Gokce, O. & Südhof, T. C. Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron 94, 611–625 (2017).
Backer, S., Sakurai, T., Grumet, M., Sotelo, C. & Bloch-Gallego, E. Nr-CAM and TAG-1 are expressed in distinct populations of developing precerebellar and cerebellar neurons. Neuroscience 113, 743–748 (2002).
Bolliger, M. F., Martinelli, D. C. & Südhof, T. C. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc. Natl Acad. Sci. USA 108, 2534–2539 (2011).
Seppälä, E. H. et al. Lgi2 truncation causes a remitting focal epilepsy in dogs. PLoS Genet. 7, e1002194 (2011).
Bravin, M., Morando, L., Vercelli, A., Rossi, F. & Strata, P. Control of spine formation by electrical activity in the adult rat cerebellum. Proc. Natl Acad. Sci. USA 96, 1704–1709 (1999).
Tyssowski, K. M. et al. Different neuronal activity patterns induce different gene expression programs. Neuron 98, 530–546 (2018).
Martinelli, D. C. et al. Expression of C1ql3 in discrete neuronal populations controls efferent synapse numbers and diverse behaviors. Neuron 91, 1034–1051 (2016).
Aimi, T., Matsuda, K. & Yuzaki, M. C1ql1–Bai3 signaling is necessary for climbing fiber synapse formation in mature Purkinje cells in coordination with neuronal activity. Mol. Brain 16, 61 (2023).
Xue, M., Atallah, B. V. & Scanziani, M. Equalizing excitation–inhibition ratios across visual cortical neurons. Nature 511, 596–600 (2014).
Gomez, A. M., Traunmüller, L. & Scheiffele, P. Neurexins: molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 22, 137–151 (2021).
Demyanenko, G. P. et al. Neural cell adhesion molecule NrCAM regulates semaphorin 3F-induced dendritic spine remodeling. J. Neurosci. 34, 11274–11287 (2014).
Han, P. et al. Cbln1 regulates axon growth and guidance in multiple neural regions. PLoS Biol. 20, e3001853 (2022).
Lanoue, V. et al. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol. Psychiatry 18, 943–950 (2013).
Fukata, Y. et al. LGI1–ADAM22–MAGUK configures transsynaptic nanoalignment for synaptic transmission and epilepsy prevention. Proc. Natl Acad. Sci. USA 118, e2022580118 (2021).
Nishida, R. et al. LOTUS, an endogenous Nogo receptor antagonist, is involved in synapse and memory formation. Sci. Rep. 11, 5085 (2021).
Petrinovic, M. M. et al. Neuronal Nogo-A negatively regulates dendritic morphology and synaptic transmission in the cerebellum. Proc. Natl Acad. Sci. USA 110, 1083–1088 (2013).
Wang, J. et al. RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development. Cell 184, 5869–5885 (2021).
Wills, Z. P. et al. The Nogo receptor family restricts synapse number in the developing hippocampus. Neuron 73, 466–481 (2012).
Thomas, R. et al. LGI1 is a nogo receptor 1 ligand that antagonizes myelin-based growth inhibition. J. Neurosci. 30, 6607–6612 (2010).
Sotelo, C. & Dusart, I. Intrinsic versus extrinsic determinants during the development of Purkinje cell dendrites. Neuroscience 162, 589–600 (2009).
Zhao, H. M., Wenthold, R. J. & Petralia, R. S. Glutamate receptor targeting to synaptic populations on Purkinje cells is developmentally regulated. J. Neurosci. 18, 5517–5528 (1998).
Cesa, R., Morando, L. & Strata, P. Glutamate receptor δ2 subunit in activity-dependent heterologous synaptic competition. J. Neurosci. 23, 2363–2370 (2003).
Matsuda, K. et al. Transsynaptic modulation of kainate receptor functions by C1q-like proteins. Neuron 90, 752–767 (2016).
Jain, S. et al. A global timing mechanism regulates cell-type-specific wiring programmes. Nature 603, 112–118 (2022).
Stroud, H. et al. An activity-mediated transition in transcription in early postnatal neurons. Neuron 107, 874–890 (2020).
Moghimyfiroozabad, S., Paul, M. A., Sigoillot, S. M. & Selimi, F. Mapping and targeting of C1ql1-expressing cells in the mouse. Sci. Rep. 13, 17563 (2023).
Hw, C. et al. Creation of a novel CRISPR-generated allele to express HA epitope-tagged C1QL1 and improved methods for its detection at synapses. FEBS Lett. 598, 2417–2437 (2024).
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Maddalo, D. et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014).
Ubina, T. et al. ExBoX—a simple Boolean exclusion strategy to drive expression in neurons. J. Cell Sci. 134, jcs257212 (2021).
Gao, Y. et al. Plug-and-play protein modification using homology-independent universal genome engineering. Neuron 103, 583–597 (2019).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Tabata, T. et al. A reliable method for culture of dissociated mouse cerebellar cells enriched for Purkinje neurons. J. Neurosci. Methods 104, 45–53 (2000).
Tervo, D. G. et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016).
Konno, K., Yamasaki, M., Miyazaki, T. & Watanabe, M. Glyoxal fixation: an approach to solve immunohistochemical problem in neuroscience research. Sci. Adv. 9, eadf7084 (2023).
Richter, K. N. et al. Glyoxal as an alternative fixative to formaldehyde in immunostaining and super‐resolution microscopy. EMBO J. 37, 139–159 (2018).
Ollion, J., Cochennec, J., Loll, F., Escudé, C. & Boudier, T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013).
De Gruijl, J. R., Bazzigaluppi, P., de Jeu, M. T. & De Zeeuw, C. I. Climbing fiber burst size and olivary sub-threshold oscillations in a network setting. PLoS Comput. Biol. 8, e1002814 (2012).
Negrello, M. et al. Quasiperiodic rhythms of the inferior olive. PLoS Comput. Biol. 15, e1006475 (2019).
Schweighofer, N., Doya, K. & Kawato, M. Electrophysiological properties of inferior olive neurons: a compartmental model. J. Neurophysiol. 82, 804–817 (1999).
Schweighofer, N. et al. Chaos may enhance information transmission in the inferior olive. Proc. Natl Acad. Sci. USA 101, 4655–4660 (2004).
Makary, S. M., Claydon, T. W., Enkvetchakul, D., Nichols, C. G. & Boyett, M. R. A difference in inward rectification and polyamine block and permeation between the Kir2.1 and Kir3.1/Kir3.4 K+ channels. J. Physiol. 568, 749–766 (2005).
Liu, G. X. et al. Comparison of cloned Kir2 channels with native inward rectifier K+ channels from guinea-pig cardiomyocytes. J. Physiol. 532, 115–126 (2001).
Picones, A., Keung, E. & Timpe, L. C. Unitary conductance variation in Kir2.1 and in cardiac inward rectifier potassium channels. Biophys. J. 81, 2035–2049 (2001).
Campbell, M. J. (ed.) Statistics at Square One 165–182 (Wiley, 2021).
Ressl, S. et al. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure 23, 688–699 (2015).
Acknowledgements
We would like to thank A. Boyreau for her help with some of the experimental procedures at the beginning of the project, Y. Dupraz for the development of tools for stereotaxic injections in neonates, F. Maloumian for her help with infographics and the personnel from the Center for Interdisciplinary Research in Biology animal and imaging facilities. We thank H. Monnet for her help in updating the plugin for VGLUT2 quantification. High-throughput qPCR was carried out on the qPCR-HD-Genomic Paris Centre platform, supported by grants from Région Ile-de-France. This work was supported by funding from Fondation pour la Recherche Médicale (Equipe FRM DEQ20150331748 to F.S.), European Research Council ERC consolidator grant (SynID 724601 to F.S.), Q-life (ANR-17-CONV-0005 to F.S. and V.H.), ANR-10-LABX-54 MEMO LIFE (to F.S.) and Sorbonne Université (ED158 to M.A.P.), Collège de France (to M.A.P.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
F.S., M.A.P. and S.M.S. conceptualized the project, developed the methodology and handled visualization. M.A.P., S.M.S., L.M., F.J.U.Q., M.D., P.M., H.W.C., E.O. and V.H. conducted the investigation. F.S. and M.A.P. secured funding. F.S. and S.M.S. provided supervision. F.S. managed project administration. F.S. wrote the original draft with contributions from S.M.S., M.A.P. and V.H. F.S., S.M.S., M.A.P., V.H. and D.C.M. reviewed and edited the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Mary Hatten and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
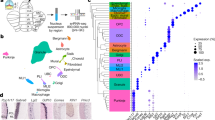
Extended Data Fig. 1 Developmental expression pattern of GC candidate genes in the cerebellar cortex.
a, The expression pattern of Cbln1 and the 15 selected GC candidate genes was assessed using high-throughput RT-qPCR on RNA extracts from the cerebellum taken at different stages of development (embryonic day, E17, postnatal days, P0, P3, P7, P14 and P21) and in adult mice. Expression levels were normalized to the Rpl13a gene and to their E17 value to capture their variation during postnatal development regardless of the individual gene’s expression level; data are presented as mean ± SEM. n = 4 animals per developmental stage and 3 animals for the adult. The data are separated into two graphs depending on the scale of dynamics of expression for clarity. b, Data from ref. 35 showing the stage of peak expression of the selected candidate genes depending on GC differentiation: GC precursors (GCPs) at P7, in newly differentiated GCs at P7, in migrating GCs at P12 and, finally, in mature GCs at P21. c, Single-cell expression data in cerebellar GCs from https://apps.kaessmannlab.org/sc-cerebellum-transcriptome/. Left: uniform manifold approximation and projection (UMAP) of mouse cerebellar cell types colored by cell differentiation state. GCP, granule cell precursor; UBCP, unipolar brush cell precursor; GC diff 1 and 2, differentiated GC1 and 2. Right: expression of Cbln1 and eleven GC DEGs candidates at different stages of GC differentiation. Dot size indicates the fraction of cells expressing a gene, color shows the mean expression level scaled by gene. d, smFISH of selected GC candidates (red) in cerebellar sections of P14 and adult mice co-stained with the nuclear stain DAPI. EGL, external granular layer; ML, molecular layer; IGL, internal granular layer. Each image corresponds to a projection of 15 images (z-stack step: 0.5 µm). Two independent experiments. Scale bars, 20 µm.
Extended Data Fig. 2 Developmental expression pattern of ION candidate genes in the inferior olive.
a, Expression patterns of C1ql1 and the 15 ION DEGs selected as candidates were assessed using high-throughput RT-qPCR on RNA extracts of the brainstem taken at different stages of development (embryonic day, E17, postnatal days, P0, P3, P7, P14 and P21) and in the adult. Expression levels were normalized to the Rpl13a gene and to their E17 value to capture their variation during postnatal development regardless of the individual gene’s expression level; data are presented as mean ± SEM. n = 4 animals per developmental stage and 3 animals for the adult. The data are separated into two graphs depending on the scale of dynamics of expression for clarity. b–d, Left: representative images from duplex smFISH experiments for C1ql1 and candidate mRNAs in coronal sections from the brainstem at P4, P14 and in adult mice. Scale bars, 150 µm. Right: the degree of correlation of expression between C1ql1 and candidate mRNAs in the brainstem was determined by computing the Pearson correlation coefficient on the whole image (coefficient >0.6 corresponding to high correlation91). Data are presented as mean ± SEM. b, Candidates highly correlated with C1ql1 at all developmental stages. Nrcam, n = 4 animals for each stage; Lgi2, adult: n = 5 animals, P14: n = 4, P4: n = 3; Crtac1, adult and P14: n = 4 animals, P4: n = 5; Sema4f, adult and P14: n = 3 animals, P4: n = 4; 2–4 independent experiments. c, Candidates highly correlated with C1ql1 from P14 to adult. Shisal1, n = 3 animals for each stage; Thy1, adult and P4: n = 3, P14: n = 4; Adam11, n = 3 for each stage; Crh: n = 3 for each stage; Gpr123, adult: n = 5 animals, P14 and P4: n = 4; 2–3 independent experiments. d, Candidates not highly correlated with C1ql1 at any stage. Tmem184b: n = 4 animals at each stage; Fstl1: n = 3 animals at each stage; Cx3cl1, adult and P14: n = 4 animals, P4: n = 3; Adam23: n = 4 animals at each stage; Cd151: n = 4 animals at each stage; Tmem179: n = 4 animals at each stage; 2–4 independent experiments.
Extended Data Fig. 3 Predicted structure and localization of C1QL1, NRCAM, LGI2 and CRTAC1 in the cerebellar cortex.
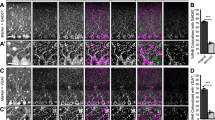
a, AlphaFold structure prediction (left; database: https://alphafold.ebi.ac.uk/) and domain architecture (right) of C1QL1, NRCAM, LGI2 and CRTAC1. See also ref. 92 for the structure of the C1QL1 gC1q domain. Ig, immunoglobulin; FN, fibronectin; TMD, transmembrane domain; LRR, leucin reach repeat; EGF, epidermal growth factor; FG-GAP, phenylalanyl-glycyl-glycyl-alanyl-prolyl. b, Localization of C1QL1, NRCAM, LGI2 and CRTAC1 proteins (green) in the cerebellar cortex at P13 was obtained using several strategies: C1QL1, anti-HA immunolabeling in cerebellar sections from a C1ql1HA knockin mouse; NRCAM, anti-HA after CRISPR–Cas9-mediated HA-tagging of endogenous NRCAM; LGI2 and CRTAC1, immunolocalization after antigen retrieval. The CF presynaptic boutons and the dendritic trees of PCs were immunostained with anti-VGLUT2 (magenta) and anti-calbindin (CaBP, blue), respectively. Each image corresponds to a projection of 5 images (z-stack step: 0.2 µm). Arrowheads show candidates labeling colocalizing with VGLUT2 clusters. 3–4 independent experiments. Scale bars, 20 µm.
Extended Data Fig. 4 CRISPR–Cas9 knockdown for C1ql1, Nrcam, Lgi2 and Crtac1.
a, Top: illustrations of the genomic regions of mouse Nrcam, Lgi2, Crtac1 and C1ql1 with the location of the sequences targeted by the CRISPR–Cas9 gRNAs. Bottom: the knockdown efficiency for Nrcam, Lgi2 and Crtac1 was measured using quantitative RT-PCR on RNA extracts from cortical cell cultures from Cas9/GFP-KI mice (Nrcam, Lgi2 and Crtac1 are detected in these cultures at DIV14). The efficiency for C1ql1 was measured using mixed cerebellar cultures from Cas9/GFP-KI-NeuroD1Cre mice expressing the CAS9 specifically in GCs precursors (C1ql1 is transiently expressed in these cultures with a peak at DIV8). Cultures were transduced at 3 or 2 days in vitro (DIV3 or 2), respectively, with AAVs driving the expression of each gRNA directed against the candidate genes or non-targeting control gRNA (CTL). Expression levels were normalized to values for the Rpl13a gene. Data are presented as mean ± SEM. n represents the independent experiments: CTL, n = 5; Crtac1 KD g1, n = 3; Crtac1 KD g2, n = 4; Lgi2 KD g1, n = 7; Lgi2 KD g2, n = 3; Nrcam KD, n = 4; C1ql1 in all conditions, n = 4. Statistics: Kruskal–Wallis and uncorrected Dunn’s test. b, Duplex smFISH experiments for GFP and each candidate gene (C1ql1, Lgi2 and Crtac1), and nuclear DAPI staining, in coronal sections from the brainstem. Top: high magnification of IONs from P21 animals with C1ql1, Lgi2 or Crtac1 KD, or non-targeting controls (CTL). Scale bars, 10 µm. Bottom: relative frequency distribution and mean integrated intensity of each candidate gene mRNA per ION under KD and CTL conditions are shown. The dashed lines represent the first quartile and second quartile (median) of the mRNA levels for each gene in CTL condition. n > 55 cells, 7–8 animals per condition, 2–3 independent experiments. Insets: data are presented as violin plots with the median and quartiles. Statistics: two-tailed nested t test. c, Distribution and mean volume of the VGLUT2 clusters quantified in GFP+ CFs after Nrcam, Lgi2 or Crtac1 KD as well as in CTL. Data are presented as violin plots with the median and quartiles. n ≥ 33 images per condition, 9–10 animals; 3–5 independent experiments. Statistics: two-tailed nested t test.
Extended Data Fig. 5 Cbln1 expression and function in IONs in the brainstem.
a, Heatmap showing the clustering of 324 DEGs according to the dynamics of their expression pattern in the brainstem between E17 and adult. Four clusters are highlighted by orange rectangles. Cbln1 is detected as an early-expressed gene. b, Top: illustration of the genomic region of mouse Cbln1 with the location of the sequence targeted by the CRISPR–Cas9 KD guide RNAs (Cbln1 KD g1 and g2) in Cbln1 exon 1 and exon 3, respectively. The location of forward (Fw) and reverse (Rev) primers used to assess deletion of the entire coding region is shown. Bottom left: efficiency of the CRISPR–Cas9 driven deletion was assessed by PCR on purified genomic DNA from DIV16 mixed cerebellar cultures (transduced at DIV2) or DIV14 neuronal cultures from the neocortex (transduced at DIV3). Wild-type fragment is expected at 2663 bp; recombined fragment at 380 bp. Right: duplex smFISH for GFP and Cbln1 mRNAs, and nuclear DAPI staining, in IONs from coronal brainstem sections of P7 mice expressing Cbln1 KD gRNAs or non-targeting CTL gRNA (GFP+). Scale bars, 10 µm. Relative frequency distribution and mean integrated intensity of Cbln1 mRNA per ION under KD and CTL conditions are shown. The dashed lines represent the first quartile and second quartile (median) of Cbln1 mRNA level in CTL condition. n ≥ 30 images per condition; 8–9 animals per condition, 2 independent experiments. Insets: data are presented as violin plots with the median and quartiles. Statistics: two-tailed nested t test. c, Distribution and mean volume of the VGLUT2 clusters were quantified in GFP+ CFs at P7 and P14 in CTL and Cbln1 KD. Data are presented as violin plots with the median and quartiles. P7 VGLUT2 cluster volume: n ≥ 29 images per condition; 8 animals, 4 independent experiments. P14 VGLUT2 cluster volume: n ≥ 33 images per condition; 8–9 animals, 3 independent experiments. Statistics: two-tailed nested t test.
Extended Data Fig. 6 Modulation of neuronal excitability by Kir2.1 expression.
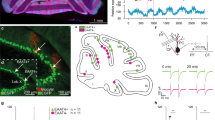
a, Neuronal cultures from the neocortex were transduced at DIV3 with a retrograde rAAV driving the expression of the Myc-tagged Kir2.1 (or nonfunctional Kir2.1 mutant as a control) and soluble eGFP under the CamKII promoter. Left: neurons were immunolabeled at DIV17 for Myc (blue) and GFP (green). Scale bars, 13 µm. Right: RNA extracts obtained from DIV14 cultures from neocortex were analyzed by quantitative RT-PCR. Gene expression levels relative to Rpl13a were normalized to the levels in Kir2.1 mutant control. Data are presented as mean ± SEM. Four independent experiments. Statistics: two-tailed Student’s t test with a null hypothesis of 1. b, In silico modeling of the effect of Kir2.1 channel expression on the activity of neurons from the inferior olive (IO). Left: representations of the discharge in neurons of the IO depending on the magnitude of the Kir2.1 conductance. Example of 10 s somatic voltage traces for different gKir conductances, ranging from gKir = 0 mS/cm2 to 0.5 mS/cm2, where, with a conservative unitary Kir2.1 channel conductance of 20 pS, 1 mS/cm2 corresponds to approximately 50 Kir2.1 channel per 100 μm2 of membrane. Right: mean firing rates and their standard deviations (for each value of gKir, n = 5 simulations of 100 s each). This modeling showed that a minimal level of Kir2.1 is needed for effective inhibition of ION activity. We selected a minimum threshold of approximately 40 Kir2.1 mRNA molecules per nucleus (as estimated by GFP mRNA intensity) to select IONs included for smFISH quantification.
Extended Data Fig. 7 Localization of GluD2 and ADGRB3/BAI3 receptors in cerebellar Purkinje cells during postnatal development.
a,b, Immunostaining of GluD2 or ADGRB3/BAI3 receptors (green), CF presynaptic boutons (anti-VGLUT2, magenta) and PCs dendritic tree (anti-CaBP, blue) in parasagittal cerebellar sections from P4 and P13 mice. Left: low magnification images correspond to the projection of 5 planes (z-stack step: 0.2 µm). Scale bars, 20 µm. Right: airyscan images corresponding to single planes. Arrowheads show receptor labeling partially colocalizing with VGLUT2 clusters. Note that at P13, some GluD2 puncta are still detectable at CF/PC synapses. Three independent experiments. Scale bars, 4 µm.
Supplementary information
Supplementary Data
Supplementary Data 1–5.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data and uncropped gel image.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Paul, M.A., Sigoillot, S.M., Marti, L. et al. Stepwise molecular specification of excitatory synapse diversity onto cerebellar Purkinje cells. Nat Neurosci 28, 308–319 (2025). https://doi.org/10.1038/s41593-024-01826-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-024-01826-w
This article is cited by
-
Structural basis of calcium-dependent C1ql1/BAI3 assemblies in synaptic connectivity
Nature Communications (2025)