Abstract
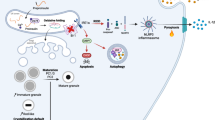
Individuals with neuropsychiatric disorders often show metabolic symptoms. However, the mechanisms underlying this co-occurrence remain unclear. Here we show that induced pluripotent stem cell-derived pancreatic islets from individuals with bipolar disorder have insulin secretion deficits caused by increased expression of RORβ, a susceptibility gene for bipolar disorder. Enhancing RORβ expression in mouse pancreatic β cells induced depression-related behaviors in the light phase and mania-like behaviors in the dark phase. Pancreatic RORβ overexpression in the light phase reduced insulin release from islets, inducing hippocampal hyperactivity and depression-like behaviors. Furthermore, this hippocampal hyperactivity in the light phase had the delayed effect of promoting insulin release in the dark phase, resulting in mania-like behaviors and hippocampal neuronal hypoactivity. Our results in mice point to a pancreas–hippocampus feedback mechanism by which metabolic and circadian factors cooperate to generate behavioral fluctuations and which may play a role in bipolar disorder.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All statistical data are available in the main text or Supplementary Information. Uncropped RT–PCR and western blot gel images are available in Supplementary Information. RNA-seq data of neurons and islet-like organoids are available in the Gene Expression Omnibus database (accession number GSE283118). Source data are available from the corresponding author.
Change history
28 August 2025
In the version of Supplementary Information initially published alongside this article, due to figure preparation errors, there were duplications in Supplementary Fig. 1k, where the sample image of HCI#4 was mistakenly replaced with that of BDI#2 and in Supplementary Fig. 3b (left), where the flow cytometry image was mistakenly duplicated from Supplementary Fig. 3a (left). The figures are now updated in the Supplementary Information.
References
Ruzickova, M., Slaney, C., Garnham, J. & Alda, M. Clinical features of bipolar disorder with and without comorbid diabetes mellitus. Can. J. Psychiatry 48, 458–461 (2003).
Hajek, T., McIntyre, R. & Alda, M. Bipolar disorders, type 2 diabetes mellitus, and the brain. Curr. Opin. Psychiatry 29, 1–6 (2016).
Wysokinski, A., Strzelecki, D. & Kloszewska, I. Levels of triglycerides, cholesterol, LDL, HDL and glucose in patients with schizophrenia, unipolar depression and bipolar disorder. Diabetes Metab. Syndr. 9, 168–176 (2015).
SayuriYamagata, A., Brietzke, E., Rosenblat, J. D., Kakar, R. & McIntyre, R. S. Medical comorbidity in bipolar disorder: the link with metabolic-inflammatory systems. J. Affect. Disord. 211, 99–106 (2017).
Calkin, C. V. et al. Insulin resistance and outcome in bipolar disorder. Br. J. Psychiatry 206, 52–57 (2015).
Steardo, L. Jr. et al. Impaired glucose metabolism in bipolar patients and response to mood stabilizer treatments. J. Affect. Disord. 245, 174–179 (2019).
Weyer, C., Bogardus, C., Mott, D. M. & Pratley, R. E. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J. Clin. Invest. 104, 787–794 (1999).
Brietzke, E. et al. Insulin dysfunction and allostatic load in bipolar disorder. Expert Rev. Neurother. 11, 1017–1028 (2011).
Talbot, F. & Nouwen, A. A review of the relationship between depression and diabetes in adults: is there a link? Diabetes Care 23, 1556–1562 (2000).
Miola, A. et al. The genetics of bipolar disorder with obesity and type 2 diabetes. J. Affect. Disord. 313, 222–231 (2022).
Derakhshan, F. & Toth, C. Insulin and the brain. Curr. Diabetes Rev. 9, 102–116 (2013).
Akhtar, A. & Sah, S. P. Insulin signaling pathway and related molecules: role in neurodegeneration and Alzheimer’s disease. Neurochem. Int. 135, 104707 (2020).
Calvo-Ochoa, E. & Arias, C. Cellular and metabolic alterations in the hippocampus caused by insulin signalling dysfunction and its association with cognitive impairment during aging and Alzheimer’s disease: studies in animal models. Diabetes Metab. Res. Rev. 31, 1–13 (2015).
Zhao, W. Q., Chen, H., Quon, M. J. & Alkon, D. L. Insulin and the insulin receptor in experimental models of learning and memory. Eur. J. Pharmacol. 490, 71–81 (2004).
Kuwabara, T. et al. Insulin biosynthesis in neuronal progenitors derived from adult hippocampus and the olfactory bulb. EMBO Mol. Med. 3, 742–754 (2011).
Ueda, H. R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002).
Ueda, H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005).
Jia, L. et al. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl Acad. Sci. USA 106, 17534–17539 (2009).
Liu, H., Aramaki, M., Fu, Y. & Forrest, D. Retinoid-related orphan receptor beta and transcriptional control of neuronal differentiation. Curr. Top. Dev. Biol. 125, 227–255 (2017).
Oishi, K., Aramaki, M. & Nakajima, K. Mutually repressive interaction between Brn1/2 and Rorb contributes to the establishment of neocortical layer 2/3 and layer 4. Proc. Natl Acad. Sci. USA 113, 3371–3376 (2016).
Zeng, H. et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149, 483–496 (2012).
Partonen, T. Clock gene variants in mood and anxiety disorders. J. Neural Transm. 119, 1133–1145 (2012).
Mansour, H. A. et al. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. 11, 701–710 (2009).
McGrath, C. L. et al. Evidence for genetic association of RORB with bipolar disorder. BMC Psychiatry 9, 70 (2009).
Lai, Y. C. et al. Investigation of associations between NR1D1, RORA and RORB genes and bipolar disorder. PLoS ONE 10, e0121245 (2015).
Baglietto, M. G. et al. RORB gene and 9q21.13 microdeletion: report on a patient with epilepsy and mild intellectual disability. Eur. J. Med. Genet. 57, 44–46 (2014).
Rudolf, G. et al. Loss of function of the retinoid-related nuclear receptor (RORB) gene and epilepsy. Eur. J. Hum. Genet. 24, 1761–1770 (2016).
Andre, E. et al. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867–3877 (1998).
Carneiro, M. et al. A loss-of-function mutation in RORB disrupts saltatorial locomotion in rabbits. PLoS Genet. 17, e1009429 (2021).
Taneera, J. et al. RORB and RORC associate with human islet dysfunction and inhibit insulin secretion in INS-1 cells. Islets 11, 10–20 (2019).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Mertens, J. et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527, 95–99 (2015).
Cogger, K. F. et al. Glycoprotein 2 is a specific cell surface marker of human pancreatic progenitors. Nat. Commun. 8, 331 (2017).
Honma, K., Hikosaka, M., Mochizuki, K. & Goda, T. Loss of circadian rhythm of circulating insulin concentration induced by high-fat diet intake is associated with disrupted rhythmic expression of circadian clock genes in the liver. Metabolism 65, 482–491 (2016).
Logan, R. W. & McClung, C. A. Animal models of bipolar mania: the past, present and future. Neuroscience 321, 163–188 (2016).
Medrihan, L. et al. Initiation of behavioral response to antidepressants by cholecystokinin neurons of the dentate gyrus. Neuron 95, 564–576 (2017).
Kheirbek, M. A. et al. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 77, 955–968 (2013).
Ramirez, S. et al. Activating positive memory engrams suppresses depression-like behaviour. Nature 522, 335–339 (2015).
Jimenez, J. C. et al. Anxiety cells in a hippocampal-hypothalamic circuit. Neuron 97, 670–683 (2018).
Milaneschi, Y., Simmons, W. K., van Rossum, E. F. C. & Penninx, B. W. Depression and obesity: evidence of shared biological mechanisms. Mol. Psychiatry 24, 18–33 (2019).
Zegarra-Valdivia, J. et al. Insulin and insulin-like growth factor-I receptors in astrocytes exert different effects on behavior and Alzheimer’s-like pathology. F1000Res 11, 663 (2022).
Kleinridders, A. et al. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc. Natl Acad. Sci. USA 112, 3463–3468 (2015).
Zhao, F., Siu, J. J., Huang, W., Askwith, C. & Cao, L. Insulin modulates excitatory synaptic transmission and synaptic plasticity in the mouse hippocampus. Neuroscience 411, 237–254 (2019).
O’Malley, D., Shanley, L. J. & Harvey, J. Insulin inhibits rat hippocampal neurones via activation of ATP-sensitive K+ and large conductance Ca2+-activated K+ channels. Neuropharmacology 44, 855–863 (2003).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Kemp, D. E. et al. Use of insulin sensitizers for the treatment of major depressive disorder: a pilot study of pioglitazone for major depression accompanied by abdominal obesity. J. Affect. Disord. 136, 1164–1173 (2012).
Kemp, D. E. et al. PPAR-gamma agonism as a modulator of mood: proof-of-concept for pioglitazone in bipolar depression. CNS Drugs 28, 571–581 (2014).
Sepanjnia, K., Modabbernia, A., Ashrafi, M., Modabbernia, M. J. & Akhondzadeh, S. Pioglitazone adjunctive therapy for moderate-to-severe major depressive disorder: randomized double-blind placebo-controlled trial. Neuropsychopharmacology 37, 2093–2100 (2012).
Zeinoddini, A. et al. Pioglitazone adjunctive therapy for depressive episode of bipolar disorder: a randomized, double-blind, placebo-controlled trial. Depress. Anxiety 32, 167–173 (2015).
Kim, B. & Feldman, E. L. Insulin resistance as a key link for the increased risk of cognitive impairment in the metabolic syndrome. Exp. Mol. Med. 47, e149 (2015).
Dalby-Brown, W. et al. Characterization of a novel high-potency positive modulator of Kv7 channels. Eur. J. Pharmacol. 709, 52–63 (2013).
Grunnet, M., Strobaek, D., Hougaard, C. & Christophersen, P. Kv7 channels as targets for anti-epileptic and psychiatric drug-development. Eur. J. Pharmacol. 726, 133–137 (2014).
Korsgaard, M. P. et al. Anxiolytic effects of Maxipost (BMS-204352) and retigabine via activation of neuronal Kv7 channels. J. Pharmacol. Exp. Ther. 314, 282–292 (2005).
Xie, X. & Hagan, R. M. Cellular and molecular actions of lamotrigine: possible mechanisms of efficacy in bipolar disorder. Neuropsychobiology 38, 119–130 (1998).
Gustavsson, N. et al. Impaired insulin secretion and glucose intolerance in synaptotagmin-7 null mutant mice. Proc. Natl Acad. Sci. USA 105, 3992–3997 (2008).
Wu, B. et al. Synaptotagmin-7 phosphorylation mediates GLP-1-dependent potentiation of insulin secretion from beta-cells. Proc. Natl Acad. Sci. USA 112, 9996–10001 (2015).
Shen, W. et al. Synaptotagmin-7 is a key factor for bipolar-like behavioral abnormalities in mice. Proc. Natl Acad. Sci. USA 117, 4392–4399 (2020).
Wang, Q. W. et al. Synaptotagmin-7 deficiency induces mania-like behavioral abnormalities through attenuating GluN2B activity. Proc. Natl Acad. Sci. USA 117, 31438–31447 (2020).
Liu, Y. N. et al. Synaptotagmin-7 deficit causes insulin hypoactivity and contributes to behavioral alterations in mice. iScience 28, 112354 (2025).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007).
Stern, S. et al. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol. Psychiatry 23, 1453–1465 (2018).
Covington, H. E. 3rd et al. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11451–11460 (2009).
Pagliuca, F. W. et al. Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439 (2014).
Mansour, A. A. et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441 (2018).
Chen, Y. et al. Synaptotagmin-1 interacts with PI(4,5)P2 to initiate synaptic vesicle docking in hippocampal neurons. Cell Rep. 34, 108842 (2021).
Zheng, Y. et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci. 21, 447–454 (2018).
Acknowledgements
We thank T. Xu (Institute of Biophysics, Chinese Academy of Science), C. Xu (Peking University) and Y. Chen (THU) for technical and material help. We thank all members of the laboratory for helpful discussions and technical assistance. This work was supported by the Beijing Natural Science Foundation (grant numbers Z210011 to J.Y. and Z200024), the National Natural Science Foundation of China (grant numbers 32371008 and 31830038), the Open Project of Collaborative Innovation Center for Language Ability of Jiangsu Province, China and the China Postdoctoral Science Foundation (grant number 2022TQ0182), and funding from Tsingha-Peking Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
Y.-N.L. Q.-W.W., C.-L.F., D.L. and X.C. conducted iPSC experiments. Y.-N.L., Q.-W.W. and B.W. conducted molecular biology and biochemistry experiments. Y.-N.L., B.W. and W. Shen conducted the animal behavioral experiments. Q.-W.W., S.-Y.L. and Y.-H.W. conducted electrophysiological experiments. L.-J.L., R.Y., C.-X.W. and L.C. conducted clinical investigations. X.-Y.S., Q.L. and S.M. conducted RNA-seq experiments. W. Shi and L.Y. provided technical assistance. Y.-N.L. and J.Y. analyzed the data. J.Y. conceived the project and designed the experiments. J.Y. wrote the paper with input from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Paul Kenny, Ignacio Torres-Aleman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Autonomous insulin signaling deficits in BDII iPSC-derived forebrain organoids.
a-c, qRT-PCR analysis showing the mRNA expression of IRS1 (a), INSR (b), and PI3K (c) in organoids before and after insulin treatment. Compared to the HCII, the BDII organoids show slightly reduced expression of these genes. In the HCII group, insulin treatment significantly decreases the PI3K expression. In the BDII group, insulin treatment increased IRS1 expression but reduced INSR expression. d, Immunoblot showing changes in InsR phosphorylation in BDII forebrain organoids. e-f, Quantitative analysis of InsR expression (e) and phosphorylation level (f). Compared to the HCII, the BDII group showed reduced levels of InsR protein expression and phosphorylation. In the HCII group, insulin treatment reduced InsR expression but enhanced InsR phosphorylation. In the BDII group, insulin treatment increased the InsR phosphorylation while the InsR protein expression remained unaffected. n = 3. One-way ANOVA with Sidak’s multiple comparisons test; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 2 RNA-seq and functional analysis of iPSC-derived islets.
a, Heatmap of selected differentially expressed genes between BDI and HCI islet-like organoids (left), and between HCII, BDII, and MDD islets (right). Values were shown as the Z score of log2(FPKM + 1) per gene. b, Table comparing the overlap of dysregulated KEGG pathways in BDI, BDII and MDD islet-like organoids. c, Venn plot showed enriched KEGG pathways based on differentially expressed genes in different groups. d-i, RNA-seq analysis showing the expression of islet marker genes, including SST (d), GCG (e), INS (f), INSR (g), NKX6.1 (h), and PDX1 (i), in the islet-like organoids of BDI/BDII/MDD subjects. n = 4 subjects (HCI), 6 (BDI), 5 (HCII/BDII/MDD). j,k, EM images (j) and quantification (k) of insulin-containing granules in mouse islet and HC/BD iPSC-derived islet-like organoids. Green arrowheads indicate insulin granules. n = 6 (mice), 9 (HC), 11 (BD). l, Mouse primary β-cells and HC iPSC-derived β-like cells showed similar levels of GSIS, n = 9 mice or HCI/II subjects. m, Sample Ca2+ imaging traces of mouse primary β-cells and HC/BDI/BDII iPSC-islets. (d-i) Two-tailed Student’s t-test; (k-l) One-way ANOVA with Sidak’s multiple comparisons test; *P < 0.05; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
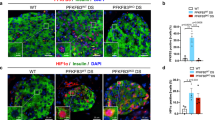
Extended Data Fig. 3 RORβ expression abnormalities in BD patient iPSC-derived islet-like organoids.
a, b, qRT-PCR analysis showing RORβ mRNA expression in the islet-like (a) and forebrain (b) organoids of each MDD donor. n = 5 subjects. c, d, RNA-seq analysis of RORβ expression in the forebrain (c) and islet-like (d) organoids of each BDI (left) and BDII/MDD (right) donor. n = 4 (HCI), 6 (BDI), 3 (HCII/BDII/MDD) subjects. Left, two-tailed Student’s t-test; right, ANOVA test. e, Immunostaining images showing RORβ expression in insulin-expressing β-cells (upper), KRT19-expressing ductal cells (middle), and PDX1-expressing β/δ/progenitor cells (lower) in BDII islet-like organoids. Scale bar, 50 µm. f–h, Quantification of RORβ expression in insulin-expressing β-cells (f; n = 38 cells for HC, 15 cells for BD), KRT19-expressing ductal cells (g; n = 79/75), and PDX1-expressing β/δ/progenitor cells (h; n = 67/67) in BDII islet-like organoids. i, ELISA analysis of insulin secretion of HCI/II iPSC-derived islets overexpressing RORβ. Cells were challenged sequentially with 2 and 20 mM glucose for three rounds, with a 30 min incubation for each concentration, followed by a depolarization with 60 mM KCl. n = 8 subjects. j, Mean GSIS level of 7 stages in i. n = 7 stages. (a-b, f-j) Two-tailed Student’s t-test. *P < 0.05; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 4 RORβ shRNA reduces RORβ expression in BD patient iPSC-derived islet-like organoids.
a, b, Immunoblot (a) and quantification (b) of RORβ protein expression in BDI/BDII islet-like organoids expressing two human RORβ shRNAs, shRNA1 and shRNA2. n = 3 subjects. c, d, ELISA analysis showing glucose-stimulated insulin secretion (GSIS) in the BDI (c) and BDII (d) islet-like organoids expressing RORβ shRNA1. Left, low/high glucose and KCl stimulated insulin release. Cells were challenged sequentially with 2 and 20 mM glucose for three rounds, with a 30-minute incubation for each concentration, followed by a depolarization with 60 mM KCl. HCI/HCII, n = 4 subjects; BDI, n = 6; BDII, n = 5. Right, Mean GSIS. n = 7 stages. (b) One-way ANOVA with Sidak’s multiple comparisons test; (c, d) Two-tailed Student’s t-test; *P < 0.05; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 5 Expression pattern of AAV-based Cre/loxP system in Ins1CreERT mice.
a, Sample immunostaining of GFP showing the expression of rAAV-EF1a-DIO-EGFP in multiple organs of tamoxifen-activated Ins1CreERT mice. Scale bar, 100 μm. b-c, Immunoblots (b) and quantification (c) of RORβ protein expression in multiple organs of sg1/2 mice. n = 4. d-e, qRT-PCR analysis of dCas9 mRNA expression in the hippocampus (d) and cortex (e) of sg1/2 mice. PC, positive control with dCas9 expression. n = 3 (PC), 7 (Ctrl), 8 (sg1/2). (c) Two-tailed Student’s t-test; (d, e) One-way ANOVA with Sidak’s multiple comparisons test; *P < 0.05; **P < 0.001; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
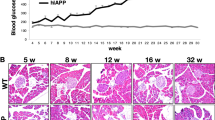
Extended Data Fig. 6 Reversed diurnal patterns of insulin function in sg1/2 mice.
a, Diurnal plasma insulin changing patterns of control (upper) and sg1/2 (lower) mice. b, Diurnal pattern of GSIS in the control and sg1/2 mice. The GSIS was determined by the ratio of glucose-stimulated versus resting condition insulin concentration. (a, b) Ctrl, from ZT5 to ZT1, n = 4/4/4/4/3/3 mice; sg1/2, n = 4/3/3/4/4/4. c-f, Immunoblot (c,e) and quantification (d,f) of insulin expression and InsR phosphorylation levels in the mouse hippocampus at the insulin peak (ZT19, 14 pm) and nadir (ZT11, 6am) of the sg1/2 mice. n = 4. g, h, Total distance of sg1/2 mice in the light (g; n = 12) and dark (h; Ctrl, n = 12; sg1/2, n = 11) periods in the open field test (OFT). i, Schematic showing injection of rAAV-EF1a-DIO-CRISPRa-mRORβ into the ventricles of Ins1CreERT mice. j, k, FST (j) and SPT (k) of vsg1/vsg2 mice in the light (white) and dark (gray) phases. n = 12. l, Immunostaining of brain sections showing that in the mice receiving rAAV-EF1a-DIO-EGFP injected into the ventricles, EGFP expression was detected in the lateral ventricles (LV) but not in the hippocampus. (a, j, k) One-way ANOVA with Sidak’s multiple comparisons test; other experiments, two-tailed Student’s t-test was used; *P < 0.05; **P < 0.001; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 7 Circadian rhythm of the sg1/2 mice.
a, Representative single-plotted actograms of voluntary wheel-running activity in light-dark (LD) and dark-dark (DD) cycles of the control and sg1/2 mice. Mice were housed in 12 L:12D for 14 days, followed by a shift to constant darkness for 7 days. White shading indicates lights on, while gray shading represents lights off. b, c, Average counts (per 6 min) of wheel-running activity in LD (b) over 14 days or in DD (c) over 7 days. The counts of the sg1/2 mice were increased compared to the control mice (n = 5). d, Average circadian free-running periods of the control and sg1/2 mice measured by chi-square periodogram in DD over 7 days (n = 5). The control and sg1/2 mice showed a similar length of period. e, Average amplitude of locomotor activity rhythm was analyzed by fast Fourier transform (FFT) in DD over 7 days. The amplitude of the sg1/2 mice was significantly increased compared to the control mice (n = 5). (b, c) Two-way ANOVA with Sidak’s multiple comparisons test; (d, e) Two-tailed Student’s t-test; *P < 0.05; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 8 Effects of manipulating islet insulin function on the brain insulin signaling and behaviors of the sg1/2 mice.
a, Plasma insulin level of sg1/2 mice during the light period. Ctrl, n = 22; sg1/2, n = 24. b, Effects of hippocampal infusion of insulin on the light-phase behaviors of the sg1/2 mice. n = 13 (FST), 11 (SPT). c, Effects of hippocampal infusion of BMS-536924 (BMS), an InsR blocker, on the light-phase behaviors of the sg1/2 mice. n = 13 (FST), 11 (SPT). d, Effects of hippocampal infusion of BMS-536924 on the plasma insulin concentration of the sg1/2 mice. n = 5 (saline), 5 (15 min), 4 (30 min). e, Immunoblot (left) and quantification of InsR expression (middle) and phosphorylation (right) in the hippocampus of the sg1/2 mice treated with repaglinide (RPG; 50 μg/kg), an anti-diabetic drug, through intraperitoneal injection (sg1/2 + RPG). n = 3. (a–c,e) Two-tailed Student’s t-test; (d) One-way ANOVA with Sidak’s multiple comparisons test; *P < 0.05; **P < 0.001; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 9 Effects of InsR knockdown on the behaviors and peripheral insulin secretion of the sg1/2 mice.
a, sg1/2 mice with InsR knockdown (KD) in the hippocampus showed slightly reduced sucrose preference (left, n = 14 (Ctrl), 13 (InsR KD)), slightly increased FST immobility time (middle, n = 13/15), and significantly decreased LDB time-in-light (right, n = 12/15) in the dark phase. Ctrl, sg1/2 mice; InsR KD, sg1/2 mice with InsR KD. b, sg1/2 mice with InsR KD in the hippocampus showed significantly reduced sucrose preference (left, n = 13/12), increased FST immobility time (middle, n = 15/15), and decreased LDB time-in-light (right, n = 14/15) in the light phase. c, ELISA analysis of plasma insulin concentration in the sg1/2 mice with InsR KD in the hippocampus. n = 5. (a, b) Two-tailed Student’s t-test; (c) One-way ANOVA with Sidak’s multiple comparison test; *P < 0.05; **P < 0.001; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Extended Data Fig. 10 Long-term effects of optical stimulations on InsR phosphorylation.
a, b, Long-term effects of a 6-hr ChR2 optical activation on the hippocampal InsR phosphorylation level of the sg1/2 (a) and control (b) mice at 6 hr after the 6-hr stimulation. D: dark phase; L: light phase. Left, immunoblot; right, quantification. ChR2, channelrhodopsin-2; NpHR, halorhodopsin. n = 4 mice for all groups. c-d, Long-term effects of a 6-hr NpHR optical suppression on the hippocampal InsR phosphorylation level of the control (c) and sg1/2 (d) mice at 6 hr after the 6-hr suppression. n = 4. Two-tailed Student’s t-test was used; *P < 0.05; error bars, s.e.m. Statistical data are listed in Supplementary Table 7.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Tables 1–7.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, YN., Wang, QW., She, XY. et al. A pancreas–hippocampus feedback mechanism regulates circadian changes in depression-related behaviors. Nat Neurosci 28, 2078–2091 (2025). https://doi.org/10.1038/s41593-025-02040-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-02040-y