Abstract
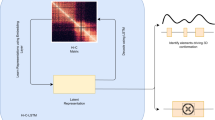
Fit-Hi-C is a programming application to compute statistical confidence estimates for Hi-C contact maps to identify significant chromatin contacts. By fitting a monotonically non-increasing spline, Fit-Hi-C captures the relationship between genomic distance and contact probability without any parametric assumption. The spline fit together with the correction of contact probabilities with respect to bin- or locus-specific biases accounts for previously characterized covariates impacting Hi-C contact counts. Fit-Hi-C is best applied for the study of mid-range (e.g., 20 kb–2 Mb for human genome) intra-chromosomal contacts; however, with the latest reimplementation, named FitHiC2, it is possible to perform genome-wide analysis for high-resolution Hi-C data, including all intra-chromosomal distances and inter-chromosomal contacts. FitHiC2 also offers a merging filter module, which eliminates indirect/bystander interactions, leading to significant reduction in the number of reported contacts without sacrificing recovery of key loops such as those between convergent CTCF binding sites. Here, we describe how to apply the FitHiC2 protocol to three use cases: (i) 5-kb resolution Hi-C data of chromosome 5 from GM12878 (a human lymphoblastoid cell line), (ii) 40-kb resolution whole-genome Hi-C data from IMR90 (human lung fibroblast), and (iii) budding yeast whole-genome Hi-C data at a single restriction cut site (EcoRI) resolution. The procedure takes ~12 h with preprocessing when all use cases are run sequentially (~4 h when run parallel). With the recent improvements in its implementation, FitHiC2 (8 processors and 16 GB memory) is also scalable to genome-wide analysis of the highest resolution (1 kb) Hi-C data available to date (~48 h with 32 GB peak memory). FitHiC2 is available through Bioconda, GitHub and the Python Package Index.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
FitHiC2 calls for different Hi-C datasets as well as processed files from published data that are used as references are provided in the Zenodo repository: https://doi.org/10.5281/zenodo.338058935.
Code availability
The source code and the documentation of FitHiC2 are publicly available through GitHub: https://github.com/ay-lab/fithic. An executable version is also provided on Code Ocean at https://codeocean.com/capsule/4528858/36. The source code is distributed under the MIT license at https://opensource.org/licenses/MIT.
References
Bickmore, W. A. The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67–84 (2013).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Kalhor, R., Tjong, H., Jayathilaka, N., Alber, F. & Chen, L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30, 90–98 (2011).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Stadhouders, R. et al. Transcription regulation by distal enhancers: who’s in the loop? Transcription 3, 181–186 (2012).
Ay, F. & Noble, W. S. Analysis methods for studying the 3D architecture of the genome. Genome Biol. 16, 183 (2015).
Lajoie, B. R., Dekker, J. & Kaplan, N. The hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75 (2015).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Yaffe, E. & Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 (2011).
Ay, F., Bailey, T. L. & Noble, W. S. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 24, 999–1011 (2014).
Bhattacharyya, S., Chandra, V., Vijayanand, P. & Ay, F. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun. 10, 4221 (2019).
Knight, P. A. & Ruiz, D. A fast algorithm for matrix balancing. IMA J. Numer. Anal. 33, 1029–1047 (2013).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Ay, F. et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 24, 974–988 (2014).
Wang, C. et al. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 25, 246–256 (2015).
Wang, M. et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587 (2017).
Ay, F. et al. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics 16, 121 (2015).
Bunnik, E. M. et al. Comparative 3D genome organization in apicomplexan parasites. Proc. Natl Acad. Sci. USA 116, 3183–3192 (2019).
Forcato, M. et al. Comparison of computational methods for Hi-C data analysis. Nat. Methods 14, 679–685 (2017).
Hwang, Y. C. et al. HIPPIE: a high-throughput identification pipeline for promoter interacting enhancer elements. Bioinformatics 31, 1290–1292 (2015).
Lun, A. T. & Smyth, G. K. diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinformatics 16, 258 (2015).
Mifsud, B. et al. GOTHiC, a probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. PLoS One 12, e0174744 (2017).
Carty, M. et al. An integrated model for detecting significant chromatin interactions from high-resolution Hi-C data. Nat. Commun. 8, 15454 (2017).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Fulco, C. P. et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019).
Chakraborty, A. & Ay, F. Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics, https://doi.org/10.1093/bioinformatics/btx664 (2017).
Dixon, J. R. et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 50, 1388–1398 (2018).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Yardimci, G. G. et al. Measuring the reproducibility and quality of Hi-C data. Genome Biol. 20, 57 (2019).
Huang, J., Marco, E., Pinello, L. & Yuan, G. C. Predicting chromatin organization using histone marks. Genome Biol. 16, 162 (2015).
Kaul, A., Bhattacharyya, S. & Ay, F. Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Zenodo, https://doi.org/10.5281/zenodo.3380589 (2019).
Kaul, A., Bhattacharyya, S. & Ay, F. Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Code Ocean, https://doi.org/10.24433/CO.5589539.v2 (2019).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Yardimci, G. G. & Noble, W. S. Software tools for visualizing Hi-C data. Genome Biol. 18, 26 (2017).
Acknowledgements
We would like to thank William S. Noble and Timothy L. Bailey for their contributions to earlier versions of Fit-Hi-C. We are also thankful to Abhijit Chakraborty for his feedback on the Fit-Hi-C package. Finally, we would like to thank all users of Fit-Hi-C/FitHiC2 who have reached out to us with their questions and valuable suggestions leading to significant improvements in the implementation and documentation. This work was funded by NIH grant R35-GM128938 to F.A.
Author information
Authors and Affiliations
Contributions
A.K. implemented the current version of FitHiC2. S.B. developed the merging filter module. A.K. and S.B. performed data analysis and wrote the manuscript under the supervision of F.A., who developed the original Fit-Hi-C code. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Ay, F. et al. Genome Res. 24, 999–1011 (2014): https://doi.org/10.1101/gr.160374.113
Sima, J. et al. Cell. 176, 816–830.e18 (2019): https://doi.org/10.1016/j.cell.2018.11.036
Bunnik, E. et al. Proc. Natl Acad. Sci. USA. 116, 3183–3192 (2019): https://doi.org/10.1073/pnas.1810815116
Zheng, Y., et al. Elife. 8, e38070 (2019): https://doi.org/10.7554/eLife.38070.001
Key data used in this protocol
Rao, S. et al. Cell. 159, 1665–1680 (2014): https://doi.org/10.1016/j.cell.2014.11.021
Quinodoz, S. et al. Cell. 174, 744–757.e24 (2018): https://doi.org/10.1016/j.cell.2018.05.024
Dixon, J. et al. Nat. Genet. 50, 1388–1398 (2018): https://doi.org/10.1038/s41588-018-0195-8
Supplementary information
Rights and permissions
About this article
Cite this article
Kaul, A., Bhattacharyya, S. & Ay, F. Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat Protoc 15, 991–1012 (2020). https://doi.org/10.1038/s41596-019-0273-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-019-0273-0
This article is cited by
-
Uncovering the multi-layer cis-regulatory landscape of rice via integrative nascent RNA analysis
Genome Biology (2025)
-
Deciphering the generation of heterogeneity in esophageal squamous cell carcinoma metastasis via single-cell multiomics analysis
Journal of Translational Medicine (2025)
-
Deciphering genetic susceptibility to clear cell renal cell carcinoma
Communications Biology (2025)
-
Multi-omic analysis of hepatocellular carcinoma reveals aberrant cis-regulatory changes and dysregulated retrotransposons with prognostic potential
Communications Biology (2025)
-
A single-cell rice atlas integrates multi-species data to reveal cis-regulatory evolution
Nature Plants (2025)