Abstract
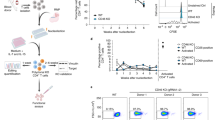
Rare cells have an important role in development and disease, and methods for isolating and studying cell subsets are therefore an essential part of biology research. Such methods traditionally rely on labeled antibodies targeted to cell surface proteins, but large public databases and sophisticated computational approaches increasingly define cell subsets on the basis of genomic, epigenomic and transcriptomic sequencing data. Methods for isolating cells on the basis of nucleic acid sequences powerfully complement these approaches by providing experimental access to cell subsets discovered in cell atlases, as well as those that cannot be otherwise isolated, including cells infected with pathogens, with specific DNA mutations or with unique transcriptional or splicing signatures. We recently developed a nucleic acid cytometry platform called ‘focused interrogation of cells by nucleic acid detection and sequencing’ (FIND-seq), capable of isolating rare cells on the basis of RNA or DNA markers, followed by bulk or single-cell transcriptomic analysis. This platform has previously been used to characterize the splicing-dependent activation of the transcription factor XBP1 in astrocytes and HIV persistence in memory CD4 T cells from people on long-term antiretroviral therapy. Here, we outline the molecular and microfluidic steps involved in performing FIND-seq, including protocol updates that allow detection and whole transcriptome sequencing of rare HIV-infected cells that harbor genetically intact virus genomes. FIND-seq requires knowledge of microfluidics, optics and molecular biology. We expect that FIND-seq, and this comprehensive protocol, will enable mechanistic studies of rare HIV+ cells, as well as other cell subsets that were previously difficult to recover and sequence.
Key points
-
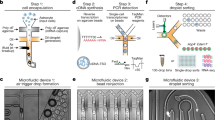
FIND-seq is a nucleic acid cytometry platform for isolating rare cells on the basis of RNA or DNA markers. This protocol describes the fabrication of three microfluidic devices and their use, with associated molecular steps, followed by the preparation of samples for bulk or single-cell transcriptomic analysis.
-
This method for isolating cells on the basis of nucleic acid sequences complements techniques that rely on labeled antibodies targeted to cell surface proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The design files for all microfluidic chips are provided in Supplementary Data 1–3. Source data are provided with this paper.
References
Siliciano, J. D. et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003).
Siliciano, J. M. & Siliciano, R. F. The remarkable stability of the latent reservoir for HIV-1 in resting memory CD4+ T cells. J. Infect. Dis. 212, 1345–1347 (2015).
Smajic, S. et al. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145, 964–978 (2022).
Xu, J. et al. Human striatal glia differentially contribute to AD- and PD-specific neurodegeneration. Nat. Aging 3, 346–365 (2023).
Bhat, S., Viswanathan, P., Chandanala, S., Prasanna, S. J. & Seetharam, R. N. Expansion and characterization of bone marrow derived human mesenchymal stromal cells in serum-free conditions. Sci. Rep. 11, 3403 (2021).
Pittenger, M. F. et al. Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen. Med. 4, 22 (2019).
Ng, Y. Y., Baert, M. R., de Haas, E. F., Pike-Overzet, K. & Staal, F. J. Isolation of human and mouse hematopoietic stem cells. Methods Mol. Biol. 506, 13–21 (2009).
Darcis, G., Berkhout, B. & Pasternak, A. O. The quest for cellular markers of HIV reservoirs: any color you like. Front. Immunol. 10, 2251 (2019).
Neidleman, J. et al. Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. eLife 9, e60933 (2020).
Lee, H. C., Chathuranga, K. & Lee, J. S. Intracellular sensing of viral genomes and viral evasion. Exp. Mol. Med. 51, 1–13 (2019).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009).
Gisler, F. M., von Kanel, T., Kraemer, R., Schaller, A. & Gallati, S. Identification of SNPs in the cystic fibrosis interactome influencing pulmonary progression in cystic fibrosis. Eur. J. Hum. Genet. 21, 397–403 (2013).
Tiscornia, G. & Mahadevan, M. S. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell 5, 959–967 (2000).
Ait Benichou, S. et al. Antisense oligonucleotides as a potential treatment for brain deficits observed in myotonic dystrophy type 1. Gene Ther. 29, 698–709 (2022).
Gierahn, T. M. et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Clark, I. C. et al. Microfluidics-free single-cell genomics with templated emulsification. Nat. Biotechnol. 41, 1557–1566 (2023).
Aldridge, S. & Teichmann, S. A. Single cell transcriptomics comes of age. Nat. Commun. 11, 4307 (2020).
Clarke, Z. A. et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat. Protoc. 16, 2749–2764 (2021).
Schreibing, F. & Kramann, R. Mapping the human kidney using single-cell genomics. Nat. Rev. Nephrol. 18, 347–360 (2022).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Mincarelli, L., Lister, A., Lipscombe, J. & Macaulay, I. C. Defining cell identity with single-cell omics. Proteomics 18, e1700312 (2018).
Sun, D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 40, 527–538 (2022).
Kharchenko, P. V. The triumphs and limitations of computational methods for scRNA-seq. Nat. Methods 18, 723–732 (2021).
Clark, I. C. et al. Identification of astrocyte regulators by nucleic acid cytometry. Nature 614, 326–333 (2023).
Clark, I. C. et al. HIV silencing and cell survival signatures in infected T cell reservoirs. Nature 614, 318–325 (2023).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Hagemann-Jensen, M., Ziegenhain, C. & Sandberg, R. Scalable single-cell RNA sequencing from full transcripts with Smart-seq3xpress. Nat. Biotechnol. 40, 1452–1457 (2022).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Shao, X. et al. scDeepSort: a pre-trained cell-type annotation method for single-cell transcriptomics using deep learning with a weighted graph neural network. Nucleic Acids Res. 49, e122 (2021).
Fa, B. et al. GapClust is a light-weight approach distinguishing rare cells from voluminous single cell expression profiles. Nat. Commun. 12, 4197 (2021).
Bej, S., Galow, A. M., David, R., Wolfien, M. & Wolkenhauer, O. Automated annotation of rare-cell types from single-cell RNA-sequencing data through synthetic oversampling. BMC Bioinformatics 22, 557 (2021).
Amamoto, R. et al. Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation. eLife 8, e51452 (2019).
Arrigucci, R. et al. FISH-Flow, a protocol for the concurrent detection of mRNA and protein in single cells using fluorescence in situ hybridization and flow cytometry. Nat. Protoc. 12, 1245–1260 (2017).
Eastburn, D. J., Sciambi, A. & Abate, A. R. Identification and genetic analysis of cancer cells with PCR-activated cell sorting. Nucleic Acids Res. 42, e128 (2014).
Clark, I. C., Thakur, R. & Abate, A. R. Concentric electrodes improve microfluidic droplet sorting. Lab Chip 18, 710–713 (2018).
Wan, L. et al. A digital microfluidic system for loop-mediated isothermal amplification and sequence specific pathogen detection. Sci. Rep. 7, 14586 (2017).
Choi, J. W., Seo, W. H., Kang, T., Kang, T. & Chung, B. G. Droplet digital recombinase polymerase amplification for multiplexed detection of human coronavirus. Lab Chip 23, 2389–2398 (2023).
Sukovich, D. J., Lance, S. T. & Abate, A. R. Sequence specific sorting of DNA molecules with FACS using 3dPCR. Sci. Rep. 7, 39385 (2017).
Hahaut, V. et al. Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq. Nat. Biotechnol. 40, 1447–1451 (2022).
Mazutis, L. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013).
Panwar, J., Autour, A. & Merten, C. A. Design and construction of a microfluidics workstation for high-throughput multi-wavelength fluorescence and transmittance activated droplet analysis and sorting. Nat. Protoc. 18, 1090–1136 (2023).
Lorenz, H. et al. High-aspect-ratio, ultrathick, negative-tone near-UV photoresist and its applications for MEMS. Sens. Actuator A Phys. 64, 33–39 (1998).
Mata, A., Fleischman, A. J. & Roy, S. Fabrication of multi-layer SU-8 microstructures. J. Micromech. Microeng. 16, 276 (2006).
Duffy, D. C., McDonald, J. C., Schueller, O. J. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Yan, Z., Clark, I. C. & Abate, A. R. Rapid encapsulation of cell and polymer solutions with bubble-triggered droplet generation. Macromol. Chem. Phys. 218, 1600297 (2017).
Bruner, K. M. et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019).
Acknowledgements
We thank members of the Clark laboratory for helpful discussions related to the development of this protocol. I.C.C. and S.W.S. were supported by K22AI152644, 1R01DA059551-01 and U54 AI170856 from the NIH.
Author information
Authors and Affiliations
Contributions
S.W.S., P.M., S.T., M.A.W. and I.C.C. designed and performed experiments or analyzed data. D.C.D., F.J.Q., E.A.B. and A.R.A. supervised the original development of FIND-seq. I.C.C. supervised the work presented in this protocol. S.W.S. and I.C.C. wrote the paper with input from their co-authors.
Corresponding author
Ethics declarations
Competing interests
I.C.C., E.A.B. and A.R.A. have filed a patent related to FIND-seq.
Peer review
Peer review information
Nature Protocols thanks Linas Mazutis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Clark, I. C. et al. Nature 614, 326–333 (2023): https://doi.org/10.1038/s41586-022-05613-0
Clark, I. C. et al. Nature 614, 318–325 (2023): https://doi.org/10.1038/s41586-022-05556-6
Supplementary information
Supplementary Information
Supplementary Figs. 1–7
Supplementary Data 1
Computer-aided design (CAD) file for the bubble-triggered microfluidic device
Supplementary Data 2
CAD file for the re-injector device
Supplementary Data 3
CAD file for the droplet sorter device
Source data
Source Data Fig. 3
Raw data for the detection efficiency and correlation curves for sample spike-in experiments
Source Data Fig. 4
Raw data for gag TaqMan assay qPCR analysis of HIV-1 provirus from sorted droplets
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shin, S.W., Mudvari, P., Thaploo, S. et al. FIND-seq: high-throughput nucleic acid cytometry for rare single-cell transcriptomics. Nat Protoc 19, 3191–3218 (2024). https://doi.org/10.1038/s41596-024-01021-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01021-y
This article is cited by
-
PURE-seq integrates FACS and PIP-seq for single-cell genomics of ultra-rare cells
Nature Communications (2026)