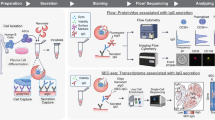
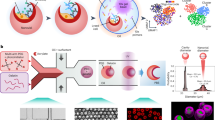
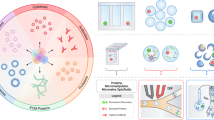
Abstract
Cells secrete numerous proteins and other biomolecules into their surroundings to achieve critical functions—from communicating with other cells to blocking the activity of pathogens. Secretion of cytokines, growth factors, extracellular vesicles and even recombinant biologic drugs defines the therapeutic potency of many cell therapies. However, gene expression states that drive specific secretory phenotypes are largely unknown. We provide a protocol that enables the secretion amount of a target protein encoded (SEC) by oligonucleotide barcodes to be linked with transcriptional sequencing (seq) for thousands of single cells. SEC-seq leverages microscale hydrogel particles called Nanovials to isolate cells and capture their secretions in close proximity, oligonucleotide-labeled antibodies to tag secretions on Nanovials and flow cytometry and single-cell RNA-sequencing (scRNA-seq) platforms for readout. Cells on Nanovials can be sorted on the basis of viability, secretion amount or other surface markers without fixation or permeabilization, and cell- and secretion-containing Nanovials are directly introduced into microfluidic droplets-in-oil emulsions for single-cell barcoding of cell transcriptomes and secretions. We have used SEC-seq to link T cell receptor sequences to the relative amount of associated cytokine secretions, surface marker gene expression with a highly secreting and potential regenerative population of mesenchymal stromal cells and the transcriptome with high immunoglobulin secretion from plasma cells. Nanovial modification and cell loading takes <4 h, and once the desired incubation time is over, staining, cell sorting and emulsion generation for scRNA-seq can also be completed in <4 h. Compared to related techniques that link secretions to a cell’s surface, SEC-seq provides a general solution across any secretion target because of the ease with which biotinylated Nanovials can be modified. By linking gene expression and secretory strength, SEC-seq can expand our understanding of cell secretion, how it is regulated and how it can be engineered to make better therapies.
Key points
-
Secretion encoded single-cell sequencing (SEC-seq) enables secretions from a single cell to be spatially associated with it in a downstream transcriptomic workflow. Microscale hydrogel particles called Nanovials isolate cells and capture secretions, which are tagged by an oligo barcode-labeled antibody for subsequent single-cell RNA-sequencing.
-
Only a few methods link secretion and transcriptomes of single cells at scale, and SEC-seq provides a broader flexibility in secretion incubation time, multiplexing and cell type compatibility.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Vogel, C. & Marcotte, E. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012).
Cheng, R. Y.-H. et al. SEC-seq: association of molecular signatures with antibody secretion in thousands of single human plasma cells. Nat. Commun. 14, 3567 (2023).
Udani, S. et al. Associating growth factor secretions and transcriptomes of single cells in nanovials using SEC-seq. Nat. Nanotechnol. 19, 354–363 (2024).
Koo, D. et al. Defining T cell receptor repertoires using nanovial-based binding and functional screening. Proc. Natl Acad. Sci. USA 121, e2320442121 (2024).
de Rutte, J. et al. Suspendable hydrogel nanovials for massively parallel single-cell functional analysis and sorting. ACS Nano 16, 7242–7257 (2022).
Lee, S., de Rutte, J., Dimatteo, R., Koo, D. & Di Carlo, D. Scalable fabrication and use of 3D structured microparticles spatially functionalized with biomolecules. ACS Nano 16, 38–49 (2022).
Ghosh, R. et al. Lab on a particle technologies. Anal. Chem. 96, 7817–7839 (2024).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Koo, D. et al. Optimizing cell therapy by sorting cells with high extracellular vesicle secretion. Nat. Commun. 15, 4870 (2024).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Miwa, H., Dimatteo, R., de Rutte, J., Ghosh, R. & Di Carlo, D. Single-cell sorting based on secreted products for functionally defined cell therapies. Microsyst. Nanoeng. 8, 84 (2022).
Healy, Z., Weinhold, K. & Murdoch, D. Transcriptional profiling of CD8+ CMV-specific T cell functional subsets obtained using a modified method for isolating high-quality RNA from fixed and permeabilized cells. Front. Immunol. 11, 1859 (2020).
Cecil, C., Nilsson, L.-A., Nygren, H., Ouchterlony, O. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J. Immunol. Methods 65, 109–121 (1983).
Ansaryan, S. et al. High-throughput spatiotemporal monitoring of single-cell secretions via plasmonic microwell arrays. Nat. Biomed. Eng. 7, 943–958 (2023).
Wu, T., Womersley, H. J., Jiehao Ray, W., Scolnick, J. & Lih Feng, C. Time-resolved assessment of single-cell protein secretion by sequencing. Nat. Methods 20, 723–734 (2023).
Xu, Y. et al. Single-cell secretion analysis via microfluidic cell membrane immunosorbent assay for immune profiling. Anal. Chem. 96, 49–58 (2024).
Dimatteo, R. & Di Carlo, D. IL-2 secretion-based sorting of single T cells using high-throughput microfluidic on-cell cytokine capture. Lab Chip 22, 1576–1583 (2022).
Rosenberg, A. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Sasagawa, Y. et al. Quartz-Seq2: a high-throughput single-cell RNA-sequencing method that effectively uses limited sequence reads. Genome Biol. 19, 29 (2018).
Macosko, E. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
de Rutte, J., Dimatteo, R., Zhu, S., Archang, M. M. & Di Carlo, D. Sorting single-cell microcarriers using commercial flow cytometers. SLAS Technol. 27, 150–159 (2022).
Koo, D., Dimatteo, R., Lee, S., de Rutte, J. & Di Carlo, D. Sorting single T cells based on secreted cytokines and surface markers using hydrogel nanovials. Preprint at https://www.biorxiv.org/content/10.1101/2022.04.28.489940v1 (2022).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Luecken, M. & Theis, F. Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Acknowledgements
We thank S. Udani for key contributions to developing the SEC-seq methodology and feedback on the manuscript. We thank D. Koo for developing the original protocol for the TCR SEC-seq workflow and for providing figures showing expected results. This project has been made possible in part by grant 2023-332386 from the Chan Zuckerberg Initiative Donor Advised Fund (CZI DAF), an advised fund of the Silicon Valley Community Foundation.
Author information
Authors and Affiliations
Contributions
J.L., S.B. and R.Y.-H.C. developed the methodology and wrote the manuscript. J.L. and S.B. contributed writing to all sections, constructed the figures and edited the manuscript. K.P. and R.G.J. contributed guidance for the writing and edited the manuscript. D.D.C. contributed writing to all sections, edited the manuscript and helped design figures.
Corresponding author
Ethics declarations
Competing interests
D.D.C. and the Regents of the University of California have financial interests in Partillion Bioscience, which sells Nanovials.
Peer review
Peer review information
Nature Protocols thanks Chia-Hung Chen and Tian Tian for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Cheng, R. Y.-H. et al. Nat. Commun. 14, 3567 (2023): https://doi.org/10.1038/s41467-023-39367-8
Udani, S. et al. Nat. Nanotechnol. 19, 354–363 (2024): https://doi.org/10.1038/s41565-023-01560-7
Koo, D. et al. Proc. Natl Acad. Sci. USA 121, e2320442121 (2024): https://doi.org/10.1073/pnas.2320442121
de Rutte, J. et al. ACS Nano 16, 7242–7257 (2022): https://doi.org/10.1021/acsnano.1c11420
Lee, S. et al. ACS Nano 16, 38–49 (2022): https://doi.org/10.1021/acsnano.1c05857
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Langerman, J., Baghdasarian, S., Cheng, R.YH. et al. Linking single-cell transcriptomes with secretion using SEC-seq. Nat Protoc 20, 2034–2055 (2025). https://doi.org/10.1038/s41596-024-01112-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01112-w
This article is cited by
-
Image-activated cell sorting
Nature Reviews Bioengineering (2025)