Abstract
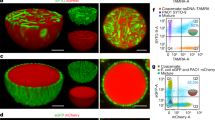
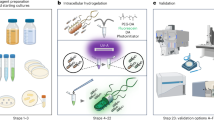
Protocell research offers diverse opportunities to understand cellular processes and the foundations of life and holds attractive potential applications across various fields. However, it is still a formidable task to construct a true-to-life synthetic cell with high organizational and functional complexity. Here we present a protocol for constructing bacteriogenic protocells by employing prokaryotes as on-site repositories of compositional, functional and structural building blocks to address this challenge. This approach is based on the capture and processing of two spatially segregated bacterial colonies within individual coacervate microdroplets to produce membrane-bounded, molecularly crowded, compositionally, structurally and functionally complex synthetic cells. The bacteriogenic protocells inherit sufficient biological components from their bacterial building units to exhibit highly integrated life-like properties, including biocatalysis, glycolysis and gene expression. The protocells can be endogenously remodeled to acquire diverse proto-organelles including a spatially partitioned nucleus-like DNA/histone-based condensate to store genetic material, membrane-bounded water vacuoles to adjust cellular osmotic pressure, a three-dimensional network of F-actin proto-cytoskeleton to support structural stability and proto-mitochondria to generate endogenous ATP as source of energy. The protocells ultimately develop a nonspherical morphology due to the continuous biogeneration of metabolic products by implanted living bacteria cells. This protocol provides a novel living material assembly strategy for the construction of functional protoliving microdevices and offers opportunities for potential applications in engineered synthetic biology and biomedicine. The protocol takes ~27 d to complete and requires expertise in microbiology, phase separation, biochemistry and molecular biology related techniques.
Key points
-
Membrane-bounded, molecularly crowded, compositionally, structurally and morphologically complex bacteriogenic protocells are constructed on the basis of the capture and on-site processing of spatially segregated bacterial colonies within individual coacervate microdroplets.
-
Bacteriogenic protocells are endogenously remodeled to acquire diverse proto-organelles and ultimately develop an amoeba-like nonspherical morphology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data discussed in this protocol are available in the supporting primary research paper17. All other data are available for research purposes from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Dzieciol, A. J. & Mann, S. Designs for life: protocell models in the laboratory. Chem. Soc. Rev. 41, 79–85 (2012).
van Stevendaal, M. H. M. E., van Hest, J. C. M. & Mason, A. F. Functional interactions between bottom-up synthetic cells and living matter for biomedical applications. ChemSystemsChem 3, e2100009 (2021).
Jeong, S., Nguyen, H. T., Kim, C. H., Ly, M. N. & Shin, K. Toward artificial cells: novel advances in energy conversion and cellular motility. Adv. Funct. Mater. 30, 1907182 (2020).
Toparlak, O. D. & Mansy, S. S. Progress in synthesizing protocells. Exp. Biol. Med. 244, 304–313 (2018).
Yewdall, N. A., Mason, A. F. & van Hest, J. C. M. The hallmarks of living systems: towards creating artificial cells. Interface Focus 8, 20180023 (2018).
Kurihara, K. et al. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 3, 775–781 (2011).
Dora Tang, T. Y. et al. Fatty acid membrane assembly on coacervate microdroplets as a step towards a hybrid protocell model. Nat. Chem. 6, 527–533 (2014).
Li, M., Harbron, R. L., Weaver, J. V. M., Binks, B. P. & Mann, S. Electrostatically gated membrane permeability in inorganic protocells. Nat. Chem. 5, 529–536 (2013).
Rodríguez-Arco, L., Li, M. & Mann, S. Phagocytosis-inspired behaviour in synthetic protocell communities of compartmentalized colloidal objects. Nat. Mater. 17, 857–863 (2017).
Qiao, Y., Li, M., Booth, R. & Mann, S. Predatory behaviour in synthetic protocell communities. Nat. Chem. 9, 110–119 (2016).
Koga, S., Williams, D. S., Perriman, A. W. & Mann, S. Peptide–nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 3, 720–724 (2011).
Huang, X. et al. Interfacial assembly of protein–polymer nano-conjugates into stimulus-responsive biomimetic protocells. Nat. Commun. 4, 2239 (2013).
Dora Tang, T. Y., van Swaay, D., deMello, A., Ross Anderson, J. L. & Mann, S. In vitro gene expression within membrane-free coacervate protocells. Chem. Commun. 51, 11429–11432 (2015).
Li, M., Green, D. C., Anderson, J. L. R., Binks, B. P. & Mann, S. In vitro gene expression and enzyme catalysis in bio-inorganic protocells. Chem. Sci. 2, 1739–1745 (2011).
Walde, P. & Ichikawa, S. Enzymes inside lipid vesicles: preparation, reactivity and applications. Biomol. Eng. 18, 143–177 (2001).
Walde, P., Goto, A., Monnard, P.-A., Wessicken, M. & Luisi, P. L. Oparin’s reactions revisited: Enzymic synthesis of Poly(adenylic acid) in micelles and self-reproducing vesicles. J Am. Chem. Soc. 116, 7541–7547 (1994).
Xu, C., Martin, N., Li, M. & Mann, S. Living material assembly of bacteriogenic protocells. Nature 609, 1029–1037 (2022).
Mann, S. Systems of creation: the emergence of life from nonliving matter. Acc. Chem. Res. 45, 2131–2141 (2012).
Xu, C., Hu, S. & Chen, X. Artificial cells: from basic science to applications. Mater. Today 19, 516–532 (2016).
Martino, C. & deMello, A. J. Droplet-based microfluidics for artificial cell generation: a brief review. Interface Focus 6, 20160011 (2016).
Solé, R. V., Munteanu, A., Rodriguez-Caso, C. & Macía, J. Synthetic protocell biology: from reproduction to computation. Philos. Trans. R. Soc. Lond. B 362, 1727–1739 (2007).
Jaffe, J. D. et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14, 1447–1461 (2004).
Thornburg, Z. R. et al. Fundamental behaviors emerge from simulations of a living minimal cell. Cell 185, 345–360.e328 (2022).
Williams, D. S. et al. Polymer/nucleotide droplets as bio-inspired functional micro-compartments. Soft Matter 8, 6004–6014 (2012).
Weibull, C. The isolation of protoplasts from Bacillus megaterium by controlled treatment with lysozyme. J. Bacteriol. 66, 688–695 (1953).
Birdsell, D. C. & Cota-Robles, E. H. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J. Bacteriol. 93, 427–437 (1967).
van den Bogaart, G., Guzman, J. V., Mika, J. T. & Poolman, B. On the mechanism of pore formation by melittin. J. Biol. Chem. 283, 33854–33857 (2008).
Pandidan, S. & Mechler, A. Nano-viscosimetry analysis of the membrane disrupting action of the bee venom peptide melittin. Sci. Rep. 9, ARTN 10841 (2019).
Oberholzer, T., Wick, R., Luisi, P. L. & Biebricher, C. K. Enzymatic RNA replication in self-reproducing vesicles—an approach to a minimal cell. Biochem. Bioph. Res. Co. 207, 250–257 (1995).
Oberholzer, T., Albrizio, M. & Luisi, P. L. Polymerase chain-reaction in liposomes. Chem. Biol. 2, 677–682 (1995).
Ishikawa, K., Sato, K., Shima, Y., Urabe, I. & Yomo, T. Expression of a cascading genetic network within liposomes. Febs. Lett. 576, 387–390 (2004).
Carlson, E. D., Gan, R., Hodgman, C. E. & Jewett, M. C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194 (2012).
Khosla, C. & Keasling, J. D. Metabolic engineering for drug discovery and development. Nat. Rev. Drug Discov. 2, 1019–1025 (2003).
Skruzny, M. et al. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl Acad. Sci. USA 109, E2533–E2542 (2012).
Emelyanov, V. V. Mitochondrial connection to the origin of the eukaryotic cell. Eur. J. Biochem. 270, 1599–1618 (2003).
Vellai, T. & Vida, G. The origin of eukaryotes: the difference between prokaryotic and eukaryotic cells. Proc. R. Soc. Lond. B 266, 1571–1577 (1999).
Drew, B. & Leeuwenburgh, C. Method for measuring ATP production in isolated mitochondria: ATP production in brain and liver mitochondria of Fischer-344 rats with age and caloric restriction. Am. J. Physiol. 285, R1259–R1267 (2003).
Acknowledgements
We thank Y. Takebayashi and J. Spencer for help with bacterial cultures; H. Sun, C. Berger-Schaffitzel and E. Bragginton for help with gel electrophoresis and western blot analysis; A. Coutable and J. L. R. Anderson for providing the plasmid pEXP5-NT/deGFP; A. Leard from the Wolfson Bioimaging Facility for help with confocal imaging; and K. Heesom from the Proteomics Facility for proteomics analysis. Special thanks to Wolfson Bioimaging Facility for providing the microscopes. C.X. was funded by China 2024 Excellent Young Scientists Fund Program (Overseas) (20241B4435) and Shanghai Pujiang Program (23PJ1406000). N.M. and S.M. were funded by the ERC Advanced Grant Scheme (EC-2016-674 ADG 740235).
Author information
Authors and Affiliations
Contributions
C.X., M.L. and S.M. conceived the protocol design. C.X. and N.M. performed the experimental procedures. C.X. and M.L. prepared the figures. C.X. and S.M. wrote the manuscript. M.L. and N.M. performed the revision. S.M. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Yan Qiao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key reference
Xu, C. et al. Nature 609, 1029–1037 (2022): https://doi.org/10.1038/s41586-022-05223-w
Supplementary information
Supplementary Information
Supplementary Figs. 1–9.
Source data
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, C., Li, M., Martin, N. et al. Construction of complex bacteriogenic protocells from living material assembly. Nat Protoc 20, 2586–2617 (2025). https://doi.org/10.1038/s41596-025-01148-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01148-6