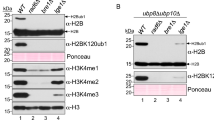
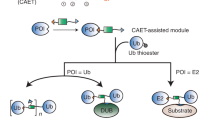
Abstract
Protein ubiquitination, a critical regulatory mechanism and post-translational modification in eukaryotic cells, involves the formation of an isopeptide bond between ubiquitin (Ub) and targeted proteins. Despite extensive investigation into the roles played by protein ubiquitination in various cellular processes, many questions remain to be answered. A major challenge in the biochemical and biophysical characterization of protein ubiquitination, along with its associated pathways and protein players, lies in the generation of ubiquitinated proteins, either in mono- or poly-ubiquitinated forms. Enzymatic and chemical strategies have been reported to address this challenge; however, there are still unmet needs for the facile generation of ubiquitinated proteins in the quantity and homogeneity required to precisely decipher the role of various protein-specific ubiquitination events. In this protocol, we provide the ubiquitin research community with a chemical ubiquitination method enabled by an α-bromoketone-mediated ligation strategy. This method can be readily adapted to generate mono- and poly-ubiquitinated proteins of interest through a cysteine introduced to replace the target lysine, with the native cysteines mutated to serine. Using proliferating cell nuclear antigen (PCNA) as an example, we present herein a detailed protocol for generating di- and tri-Ub PCNA that contains a photo-activatable cross-linker for capturing potential reader proteins. The thioether-mediated protein ligation and purification typically takes 2–3 weeks. An important feature of our ubiquitination strategy is the ability to introduce a Michael-acceptor warhead to the linkage, allowing the generation of activity-based probes for deubiquitinases and ubiquitin-carrying enzymes such as HECT and RBR E3 ubiquitin ligases and E2 enzymes. As such, our method is highly versatile and can be readily adapted to investigate the readers and erasers of many proteins that undergo reversible ubiquitination.
Key points
-
Studying the role of protein ubiquitination requires well-characterized ubiquitinated protein derivatives. This protocol describes a semisynthetic chemical strategy that uses an α-bromoketone non-hydrolyzable linker (NHL) to connect ubiquitin to proteins of interest (POI).
-
An NHL variant containing a Michael acceptor can be used to trap deubiquitinases (DUBs) and ubiquitin-carrying enzymes. This protocol describes how to prepare the NHL and starting materials and perform reactions with POIs and ubiquitin species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this work are provided within the paper. The raw data and related files not included in the current paper can be requested from the corresponding author. Source data are provided with this paper.
References
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Clague, M. J., Heride, C. & Urbé, S. The demographics of the ubiquitin system. Trends Cell Biol. 25, 417–426 (2015).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Deol, K. K., Lorenz, S. & Strieter, E. R. Enzymatic logic of ubiquitin chain assembly. Front. Physiol. 10, 835 (2019).
Sun, M. & Zhang, X. Current methodologies in protein ubiquitination characterization: from ubiquitinated protein to ubiquitin chain architecture. Cell Biosci. 12, 126 (2022).
Faggiano, S. & Pastore, A. The challenge of producing ubiquitinated proteins for structural studies. Cells 3, 639–656 (2014).
Chatterjee, C., McGinty, R. K., Pellois, J. P. & Muir, T. W. Auxiliary-mediated site-specific peptide ubiquitylation. Angew. Chem. Int. Ed. Engl. 46, 2814–2818 (2007).
Yang, R., Pasunooti, K. K., Li, F., Liu, X. W. & Liu, C. F. Dual native chemical ligation at lysine. J. Am. Chem. Soc. 131, 13592–13593 (2009).
Ajish Kumar, K. S., Haj-Yahya, M., Olschewski, D., Lashuel, H. A. & Brik, A. Highly efficient and chemoselective peptide ubiquitylation. Angew. Chem. Int. Ed. Engl. 48, 8090–8094 (2009).
Haj-Yahya, M., Ajish Kumar, K. S., Erlich, L. A. & Brik, A. Protecting group variations of delta-mercaptolysine useful in chemical ubiquitylation. Biopolymers 94, 504–510 (2010).
Virdee, S. et al. Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 133, 10708–10711 (2011).
Weller, C. E., Huang, W. & Chatterjee, C. Facile synthesis of native and protease-resistant ubiquitylated peptides. Chembiochem 15, 1263–1267 (2014).
Weller, C. E. et al. Aromatic thiol-mediated cleavage of N-O bonds enables chemical ubiquitylation of folded proteins. Nat. Commun. 7, 12979 (2016).
Pan, M. et al. Quasi-racemic X-ray structures of K27-linked ubiquitin chains prepared by total chemical synthesis. J. Am. Chem. Soc. 138, 7429–7435 (2016).
Burlina, F. et al. Auxiliary-assisted chemical ubiquitylation of NEMO and linear extension by HOIP. Commun. Chem. 2, 111 (2019).
Sun, H. et al. Diverse fate of ubiquitin chain moieties: the proximal is degraded with the target, and the distal protects the proximal from removal and recycles. Proc. Natl. Acad. Sci. Usa. 116, 7805–7812 (2019).
Layfield, R. et al. Chemically synthesized ubiquitin extension proteins detect distinct catalytic capacities of deubiquitinating enzymes. Anal. Biochem. 274, 40–49 (1999).
Kumar, K. S., Spasser, L., Ohayon, S., Erlich, L. A. & Brik, A. Expeditious chemical synthesis of ubiquitinated peptides employing orthogonal protection and native chemical ligation. Bioconjug. Chem. 22, 137–143 (2011).
Cardella, D., Tsai, Y.-H. & Luk, L. Y. P. Towards the use of an amino acid cleavable linker for solid-phase chemical synthesis of peptides and proteins. Org. Biomol. Chem. 21, 966–969 (2023).
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Castañeda, C. et al. Nonenzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J. Am. Chem. Soc. 133, 17855–17868 (2011).
Yin, L., Krantz, B., Russell, N. S., Deshpande, S. & Wilkinson, K. D. Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochemistry 39, 10001–10010 (2000).
Lewis, Y. E., Abeywardana, T., Lin, Y. H., Galesic, A. & Pratt, M. R. Synthesis of a bis-thio-acetone (BTA) analogue of the lysine isopeptide bond and its application to investigate the effects of ubiquitination and SUMOylation on α-synuclein aggregation and toxicity. ACS Chem. Biol. 11, 931–942 (2016).
Yang, K. et al. Chemical protein ubiquitylation with preservation of the native cysteine residues. Chembiochem 17, 995–998 (2016).
Morgan, M. T. et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016).
Chen, J., Ai, Y., Wang, J., Haracska, L. & Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 6, 270–272 (2010).
Chatterjee, C., McGinty, R. K., Fierz, B. & Muir, T. W. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 6, 267–269 (2010).
Meier, F. et al. Semisynthetic, site-specific ubiquitin modification of alpha-synuclein reveals differential effects on aggregation. J. Am. Chem. Soc. 134, 5468–5471 (2012).
Hemantha, H. P. et al. Nonenzymatic polyubiquitination of expressed proteins. J. Am. Chem. Soc. 136, 2665–2673 (2014).
Yang, K., Gong, P., Gokhale, P. & Zhuang, Z. Chemical protein polyubiquitination reveals the role of a noncanonical polyubiquitin chain in DNA damage tolerance. ACS Chem. Biol. 9, 1685–1691 (2014).
Weikart, N. D. & Mootz, H. D. Generation of site-specific and enzymatically stable conjugates of recombinant proteins with ubiquitin-like modifiers by the Cu(I)-catalyzed azide-alkyne cycloaddition. Chembiochem 11, 774–777 (2010).
Eger, S., Scheffner, M., Marx, A. & Rubini, M. Synthesis of defined ubiquitin dimers. J. Am. Chem. Soc. 132, 16337–16339 (2010).
Eger, S. et al. Generation of a mono-ubiquitinated PCNA mimic by click chemistry. Chembiochem 12, 2807–2812 (2011).
Trang, V. H. et al. Nonenzymatic polymerization of ubiquitin: single-step synthesis and isolation of discrete ubiquitin oligomers. Angew. Chem. Int. Ed. Engl. 51, 13085–13088 (2012).
Valkevich, E. M. et al. Forging isopeptide bonds using thiol-ene chemistry: site-specific coupling of ubiquitin molecules for studying the activity of isopeptidases. J. Am. Chem. Soc. 134, 6916–6919 (2012).
Shanmugham, A. et al. Nonhydrolyzable ubiquitin-isopeptide isosteres as deubiquitinating enzyme probes. J. Am. Chem. Soc. 132, 8834–8835 (2010).
Stanley, M. & Virdee, S. Genetically directed production of recombinant, isosteric and nonhydrolysable ubiquitin conjugates. Chembiochem 17, 1472–1480 (2016).
Singh, S. K. et al. Synthetic uncleavable ubiquitinated proteins dissect proteasome deubiquitination and degradation, and highlight distinctive fate of tetraubiquitin. J. Am. Chem. Soc. 138, 16004–16015 (2016).
Bhat, S. et al. Hydrazide mimics for protein lysine acylation to assess nucleosome dynamics and deubiquitinase action. J. Am. Chem. Soc. 140, 9478–9485 (2018).
Dixon, E. K., Castañeda, C. A., Kashyap, T. R., Wang, Y. & Fushman, D. Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies. Bioorg. Med. Chem. 21, 3421–3429 (2013).
Meledin, R., Mali, S. M., Singh, S. K. & Brik, A. Protein ubiquitination via dehydroalanine: development and insights into the diastereoselective 1,4-addition step. Org. Biomol. Chem. 14, 4817–4823 (2016).
Baumann, A. L. et al. Chemically induced vinylphosphonothiolate electrophiles for thiol-thiol bioconjugations. J. Am. Chem. Soc. 142, 9544–9552 (2020).
Fottner, M. et al. Site-specific ubiquitylation and SUMOylation using genetic-code expansion and sortase. Nat. Chem. Biol. 15, 276–284 (2019).
Raniszewski, N. R., Beyer, J. N., Noel, M. I. & Burslem, G. M. Sortase mediated protein ubiquitination with defined chain length and topology. RSC Chem. Biol. 5, 321–327 (2024).
Chu, G. C. et al. Cysteine-aminoethylation-assisted chemical ubiquitination of recombinant histones. J. Am. Chem. Soc. 141, 3654–3663 (2019).
Zheng, Q. et al. An E1-catalyzed chemoenzymatic strategy to isopeptide-N-ethylated deubiquitylase-resistant ubiquitin probes. Angew. Chem. Int. Ed. Engl. 59, 13496–13501 (2020).
Gui, W., Davidson, G. A. & Zhuang, Z. Chemical methods for protein site-specific ubiquitination. RSC Chem. Biol. 2, 450–467 (2021).
Kriegesmann, J. & Brik, A. Synthesis of ubiquitinated proteins for biochemical and functional analysis. Chem. Sci. 14, 10025–10040 (2023).
McGinty, R. K., Kim, J., Chatterjee, C., Roeder, R. G. & Muir, T. W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 (2008).
Hejjaoui, M., Haj-Yahya, M., Kumar, K. S., Brik, A. & Lashuel, H. A. Towards elucidation of the role of ubiquitination in the pathogenesis of Parkinson’s disease with semisynthetic ubiquitinated α-synuclein. Angew. Chem. Int. Ed. Engl. 50, 405–409 (2011).
González-Magaña, A. & Blanco, F. J. Human PCNA structure, function and interactions. Biomolecules 10, 570 (2020).
Powers, K. T. & Washington, M. T. Eukaryotic translesion synthesis: choosing the right tool for the job. DNA Repair (Amst.) 71, 127–134 (2018).
Xu, X. et al. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev. Mutat. Res. 764, 43–50 (2015).
Das-Bradoo, S. et al. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat. Cell Biol. 12, 74–79 (2010). sup pp 1–20.
Yuan, J., Ghosal, G. & Chen, J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47, 410–421 (2012).
Weston, R., Peeters, H. & Ahel, D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26, 1558–1572 (2012).
Ciccia, A. et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell 47, 396–409 (2012).
Saugar, I., Parker, J. L., Zhao, S. & Ulrich, H. D. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res. 40, 245–257 (2012).
Gong, P. et al. Activity-based ubiquitin-protein probes reveal target protein specificity of deubiquitinating enzymes. Chem. Sci. 9, 7859–7865 (2018).
Shen, S., Davidson, G. A., Yang, K. & Zhuang, Z. Photo-activatable Ub-PCNA probes reveal new structural features of the Saccharomyces cerevisiae Poleta/PCNA complex. Nucleic Acids Res. 49, 9374–9388 (2021).
Li, G., Liang, Q., Gong, P., Tencer, A. H. & Zhuang, Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem. Commun. 50, 216–218 (2014).
Paudel, P. et al. Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X. Proc. Natl. Acad. Sci. Usa. 116, 7288–7297 (2019).
Paudel, P., Banos, C. M., Liu, Y. & Zhuang, Z. Triubiquitin probes for identification of reader and eraser proteins of branched polyubiquitin chains. ACS Chem. Biol. 18, 837–847 (2023).
Komander, D., Clague, M. J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Bello, A. I. et al. Deubiquitinases in neurodegeneration. Cells 11, 556 (2022).
He, M. et al. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 6, 62 (2016).
Hehl, L. A. et al. Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5. Nat. Chem. Biol. 20, 190–200 (2024).
Horn-Ghetko, D. et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature 590, 671–676 (2021).
McGouran, J. F., Gaertner, S. R., Altun, M., Kramer, H. B. & Kessler, B. M. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem. Biol. 20, 1447–1455 (2013).
Haj-Yahya, N. et al. Dehydroalanine-based diubiquitin activity probes. Org. Lett. 16, 540–543 (2014).
Mulder, M. P., El Oualid, F., ter Beek, J. & Ovaa, H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. Chembiochem 15, 946–949 (2014).
Meledin, R., Mali, S. M., Kleifeld, O. & Brik, A. Activity-based probes developed by applying a sequential dehydroalanine formation strategy to expressed proteins reveal a potential α-globin-modulating deubiquitinase. Angew. Chem. Int. Ed. Engl. 57, 5645–5649 (2018).
Pao, K. C. et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat. Chem. Biol. 12, 324–331 (2016).
Pao, K. C. et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556, 381–385 (2018).
Xu, L. et al. An activity-based probe developed by a sequential dehydroalanine formation strategy targets HECT E3 ubiquitin ligases. Chem. Commun. (Camb.) 55, 7109–7112 (2019).
Nguyen, D. P. et al. Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc. 136, 2240–2243 (2014).
Chojnacki, M. et al. Polyubiquitin-photoactivatable crosslinking reagents for mapping ubiquitin interactome identify Rpn1 as a proteasome ubiquitin-associating subunit. Cell Chem. Biol. 24, 443–457.e6 (2017).
Liang, J. et al. Chemical synthesis of diubiquitin-based photoaffinity probes for selectively profiling ubiquitin-binding proteins. Angew. Chem. Int. Ed. Engl. 56, 2744–2748 (2017).
Tan, X. D. et al. A diubiquitin-based photoaffinity probe for profiling K27-linkage targeting deubiquitinases. Chem. Commun. (Camb.) 53, 10208–10211 (2017).
Mathur, S., Fletcher, A. J., Branigan, E., Hay, R. T. & Virdee, S. Photocrosslinking activity-based probes for ubiquitin RING E3 ligases. Cell Chem. Biol. 27, 74–82.e6 (2020).
Fan, L., Bi, T., Wang, L. & Xiao, W. DNA-damage tolerance through PCNA ubiquitination and sumoylation. Biochem. J. 477, 2655–2677 (2020).
Choe, K. N. & Moldovan, G.-L. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell 65, 380–392 (2017).
Zheng, Q. et al. A bifunctional molecule-assisted synthesis of mimics for use in probing the ubiquitination system. Nat. Protoc. 18, 530–554 (2023).
Liwocha, J. et al. Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases. Nat. Struct. Mol. Biol. 31, 378–389 (2024).
Li, J. et al. Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting. Mol. Cell 84, 1304–1320.e16 (2024).
Acknowledgements
This work was supported by National Institutes of Health grants R01GM129468, R35GM152011 and R21AG077189 to Z.Z. and National Institute of General Medical Sciences grants P30 GM110758 and P20 GM104316.
Author information
Authors and Affiliations
Contributions
G.A.D. performed the ubiquitin NHL modifications and ligation to PCNA. Z.M. performed the ubiquitin MAL modifications. A.R.S. performed the synthesis of the linker molecules. G.A.D., Z.M., A.R.S. and Z.Z. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Z.Z. has received a patent (US 9,605,297 B2) on the thioether linker-mediated protein ligation method.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Li, G. et al. Chem. Commun. 50, 216–218 (2014): https://doi.org/10.1039/c3cc47382a
Yang, K. et al. ChemBioChem 17, 995–998 (2016): https://doi.org/10.1002/cbic.201600042
Gong, P. et al. Chem. Sci. 9, 7859–7865 (2018): https://doi.org/10.1039/c8sc01573b
Paudel, P. et al. Proc. Natl. Acad. Sci. USA 116, 7288–7297 (2019): https://doi.org/10.1073/pnas.1815027116
Shen, S. et al. Nucleic Acids Res. 49, 9374–9388 (2021): https://doi.org/10.1093/nar/gkab646
Supplementary information
Supplementary Information
Supplementary Figures 1–7
Source data
Source Data Figs. 2–4
Unprocessed gels
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Davidson, G.A., Moafian, Z., Sensi, A.R. et al. Thioether-mediated protein ubiquitination in constructing affinity- and activity-based ubiquitinated protein probes. Nat Protoc 20, 3239–3269 (2025). https://doi.org/10.1038/s41596-025-01162-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01162-8