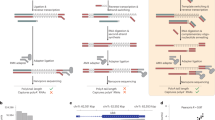
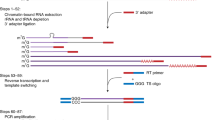
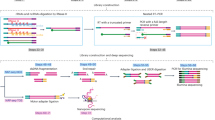
Abstract
RNA polyadenylation is crucial for RNA maturation, stability and function, with poly(A) tail lengths significantly influencing mRNA translation, efficiency and decay. Here, we provide a step-by-step protocol to perform Nanopore 3′ end-capture sequencing (Nano3P-seq), a nanopore-based cDNA sequencing method to simultaneously capture RNA abundances and tail-composition and tail-length estimates at single-molecule resolution. Taking advantage of a template switching–based protocol, Nano3P-seq can sequence any RNA-derived molecule from its 3′ end, regardless of its polyadenylation status, without the need for PCR amplification or RNA adapter ligation. We provide an updated Nano3P-seq protocol that is compatible with R10.4 flow cells, as well as compatible software for poly(A) tail length and content prediction, which we term ‘PolyTailor’. We demonstrate that PolyTailor provides accurate estimates of transcript abundances and tail lengths and composition, while capturing both coding and noncoding RNA biotypes, including mRNAs, small nucleolar RNAs and ribosomal RNAs. Nano3P-seq can be applied to RNA samples prepared by using different methods (e.g., poly(A)-selected, ribodepleted or total RNA) and can be completed in 1 day. The protocol requires experience in molecular biology techniques and nanopore sequencing library preparation and basic knowledge of Linux Bash syntax and R programming. This protocol makes Nano3P-seq accessible and easy to implement by future users aiming to study the tail dynamics and heterogeneity of both coding and noncoding transcriptomes in a comprehensive and reproducible manner.
Key points
-
Nano3P-seq uses nanopore-based cDNA sequencing to characterize multiple coding and noncoding RNA biotypes from RNA prepared by using various methods, including poly(A)-selected, ribodepleted or total RNA.
-
The protocol is compatible with the latest R10 sequencing chemistry from Oxford Nanopore Technologies and provides a complementary software package, PolyTailor, to estimate RNA abundance and tail length and composition at the single-molecule level.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data used in this protocol have been deposited in the European Nucleotide Archive at the European Molecular Biology Laboratory-European Bioinformatics Institute under accession number PRJEB80101. A subset of the Nano3P-seq reads used in this manuscript (and their respective reference files) can be downloaded from the Novoa Lab’s public repository for pipeline testing at https://public-docs.crg.es/enovoa/public/lpryszcz/src/polyTailor/test/.
Code availability
The PolyTailor software used for Nano3P-seq analysis, including test data, has been made publicly available in GitHub at https://github.com/novoalab/polyTailor32.
References
Weill, L., Belloc, E., Bava, F.-A. & Méndez, R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 19, 577–585 (2012).
Passmore, L. A. & Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 23, 93–106 (2021).
Brouze, A., Krawczyk, P. S., Dziembowski, A. & Mroczek, S. Measuring the tail: methods for poly(A) tail profiling. Wiley Interdiscip. Rev. RNA 14, e1737 (2023).
Liu, J. & Lu, F. Beyond simple tails: poly(A) tail-mediated RNA epigenetic regulation. Trends Biochem. Sci. 49, 846–858 (2024).
Lucas, M. C. & Novoa, E. M. Long-read sequencing in the era of epigenomics and epitranscriptomics. Nat. Methods 20, 25–29 (2023).
Jain, M., Olsen, H. E., Paten, B. & Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 (2016).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Begik, O. et al. Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore cDNA sequencing. Nat. Methods 20, 75–85 (2023).
Delgado-Tejedor, A. et al. Native RNA nanopore sequencing reveals antibiotic-induced loss of rRNA modifications in the A- and P-sites. Nat. Commun. 15, 10054 (2024).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
Adiconis, X. et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods 10, 623–629 (2013).
Zhao, L. et al. Analysis of transcriptome and epitranscriptome in plants using PacBio Iso-Seq and nanopore-based direct RNA sequencing. Front. Genet. 10, 253 (2019).
Oikonomopoulos, S. et al. Methodologies for transcript profiling using long-read technologies. Front. Genet. 11, 606 (2020).
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Au, K. F. et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc. Natl. Acad. Sci. USA 110, E4821–E4830 (2013).
Hardwick, S. A. et al. Spliced synthetic genes as internal controls in RNA sequencing experiments. Nat. Methods 13, 792–798 (2016).
Begik, O. et al. Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore sequencing. Nat. Methods 20, 75–85 (2023).
Legnini, I., Alles, J., Karaiskos, N., Ayoub, S. & Rajewsky, N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods 16, 879–886 (2019).
Liu, Y., Nie, H., Liu, H. & Lu, F. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat. Commun. 10, 5292 (2019).
Liu, Y., Nie, H., Zhang, Y., Lu, F. & Wang, J. Comprehensive analysis of mRNA poly(A) tails by PAIso-seq2. Sci. China Life Sci. 66, 187–190 (2023).
Long, Y., Jia, J., Mo, W., Jin, X. & Zhai, J. FLEP-seq: simultaneous detection of RNA polymerase II position, splicing status, polyadenylation site and poly(A) tail length at genome-wide scale by single-molecule nascent RNA sequencing. Nat. Protoc. 16, 4355–4381 (2021).
Jia, J. et al. An atlas of plant full-length RNA reveals tissue-specific and monocots-dicots conserved regulation of poly(A) tail length. Nat. Plants 8, 1118–1126 (2022).
Gumińska, N. et al. LRB-IIMCB/ninetails: v.1.0.2_manuscript. Zenodo https://zenodo.org/records/13837855 (2024).
Nottingham, R. M. et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22, 597–613 (2016).
Wu, D. C. & Lambowitz, A. M. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Sci. Rep. 7, 8421 (2017).
Xu, H., Yao, J., Wu, D. C. & Lambowitz, A. M. Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Sci. Rep. 9, 7953 (2019).
Mohr, S. et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19, 958–970 (2013).
Lentzsch, A. M., Yao, J., Russell, R. & Lambowitz, A. M. Template-switching mechanism of a group II intron-encoded reverse transcriptase and its implications for biological function and RNA-Seq. J. Biol. Chem. 294, 19764–19784 (2019).
Behrens, A., Rodschinka, G. & Nedialkova, D. D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815.e7 (2021).
Prjibelski, A. D. et al. Accurate isoform discovery with IsoQuant using long reads. Nat. Biotechnol. 41, 915–918 (2023).
Xu, H., Nottingham, R. M. & Lambowitz, A. M. TGIRT-seq protocol for the comprehensive profiling of coding and non-coding RNA biotypes in cellular, extracellular vesicle, and plasma RNAs. Bio Protoc. 11, 23 (2021).
Begik, O., Pryszcz, L. P., Niazi, A. M., Valen, E. & Novoa, E. M. Nano3P-seq: charting the coding and non-coding transcriptome at single-molecule resolution. Zenodo https://zenodo.org/records/14082765 (2024).
Acknowledgements
This work was supported by the Spanish Ministry of Science, Innovation and Universities (MCIN/AEI/10.13039/501100011033/FEDER, UE) (PID2021-128193NB-100 to E.M.N.). This project received funding from the European Union’s Horizon Europe under the grant agreement No. 101042103 (to E.M.N.). The views and opinions expressed are, however, those of the author(s) only and do not necessarily reflect those of the European Union. Neither the European Union nor the granting authority can be held responsible for these opinions. O.B. was supported by CRG Proof-Of-Concept funding. We acknowledge support from the Spanish Ministry of Science and Innovation through the Centro de Excelencia Severo Ochoa (CEX2020-001049-S, MCIN/AEI /10.13039/501100011033), the Generalitat de Catalunya through the CERCA programme and the EMBL partnership. We are grateful to the CRG Core Technologies Programme for their support and assistance in this work. Figures were drawn by using BioRender.
Author information
Authors and Affiliations
Contributions
O.B. adapted the experimental protocol for Nano3P-seq for R10 flow cells. L.P.P. developed the PolyTailor software. O.B., A.M.N., E.V. and E.M.N. troubleshooted the tail-length prediction tool development. O.B. built the figures. E.M.N. supervised the project. O.B., L.P. and E.M.N. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.M.N. is a member of the Scientific Advisory Board of IMMAGINA Biotech. O.B. and E.M.N. have received travel bursaries from ONT to present their work at conferences.
Peer review
Peer review information
Nature Protocols thanks Bin Tian, Falong Lu, Jingwen Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Begik, O. et al. Nat. Methods 20, 75–85 (2023): https://doi.org/10.1038/s41592-022-01714-w
Delgado-Tejedor, A. et al. Nat. Commun. 15, 10054 (2024): https://doi.org/10.1038/s41467-024-54368-x
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Begik, O., Pryszcz, L.P., Niazi, A.M. et al. Nano3P-seq: charting the coding and noncoding transcriptome at single-molecule resolution. Nat Protoc 20, 3607–3628 (2025). https://doi.org/10.1038/s41596-025-01205-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01205-0