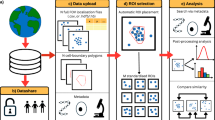
Abstract
Super-resolution microscopy has revolutionized the ability to investigate biological structures and processes, which are now accessible at nanoscale resolution. Recent advances in single-particle tracking (SPT) approaches have enabled researchers to study the intermolecular dynamics of individual proteins within their native environments in live cells. Fluorescent intrabody localization microscopy expands on existing SPT approaches such as SPT photoactivated localization microscopy by granting access to the nanoclustering dynamics of intracellular endogenous proteins through the use of single-domain nanobodies that can also differentiate between the conformational states of proteins. Here we detail how to perform single-molecule imaging of expressed proteins and nanobodies raised against endogenous proteins. We provide a streamlined analytical pipeline utilizing newly established clustering algorithms for extracting meaningful biological information. Nanoclustering analysis using spatiotemporal indexing is an open-source program with a user interface that enables the extraction of a range of dynamic nanoclustering metrics, including spatial and temporal information, from SPT data. This Protocol combines these single-molecule tracking and spatiotemporal clustering approaches into a comprehensive guide for researchers to achieve the precise localization of expressed and endogenous proteins and the characterization of their conformation-specific clustering behavior within subcellular compartments at nanoscale resolution. The procedure requires 2–4 d and is suitable for users with some prior experience in super-resolution microscopy and microscopy data analysis.
Key points
-
FiLM and sptPALM enable the single-particle tracking of endogenous and expressed proteins, either directly or by proxy, respectively, in living cells. NASTIC and segNASTIC capture the spatial and temporal dynamics of tracked proteins in clusters using single-particle tracking data to characterize their nanoscale organization in living cells.
-
FiLM uses unique nanobodies and biosensors to target and track specific protein conformation subpopulations, and NASTIC interrogates their mobility and dynamic spatiotemporal clustering behavior.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data presented in ‘Anticipated results’ were acquired from previous journal articles4,5. They are available via https://doi.org/10.14264/03a862c and https://doi.org/10.1038/s41467-023-38866-y or upon request from the respective corresponding authors.
Code availability
NASTIC software, File Converter and Wrangler GUI, and all associated codes and user manuals are available via GitHub at https://github.com/tristanwallis/smlm_clustering.
References
Gormal, R. S. & Meunier, F. A. Nanoscale organization of the pre-synapse: tracking the neurotransmitter release machinery. Curr. Opin. Neurobiol. 75, 102576 (2022).
Gormal, R. S., Martinez-Marmol, R., Brooks, A. J. & Meunier, F. A. Location, location, location: protein kinase nanoclustering for optimised signalling output. eLife https://doi.org/10.7554/eLife.93902 (2024).
Harding, A. S. & Hancock, J. F. Using plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 18, 364–371 (2008).
Gormal, R. S. et al. Modular transient nanoclustering of activated β2-adrenergic receptors revealed by single-molecule tracking of conformation-specific nanobodies. Proc. Natl Acad. Sci. USA 117, 30476–30487 (2020).
Wallis, T. P. et al. Super-resolved trajectory-derived nanoclustering analysis using spatiotemporal indexing. Nat. Commun. 14, 3353 (2023).
Manley, S. et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008).
Chhabra, Y. et al. Tyrosine kinases compete for growth hormone receptor binding and regulate receptor mobility and degradation. Cell Rep. 42, 112490 (2023).
Martinez-Marmol, R. et al. Fyn nanoclustering requires switching to an open conformation and is enhanced by FTLD-Tau biomolecular condensates. Mol. Psychiatry 28, 946–962 (2023).
Padmanabhan, P., Martinez-Marmol, R., Xia, D., Gotz, J. & Meunier, F. A. Frontotemporal dementia mutant Tau promotes aberrant Fyn nanoclustering in hippocampal dendritic spines. eLife https://doi.org/10.7554/eLife.45040 (2019).
Griffié, J. et al. Dynamic Bayesian cluster analysis of live-cell single molecule localization microscopy datasets. Small Methods 2, ARTN 1800008 (2018).
Guttman, A. R-trees: a dynamic index structure for spatial searching. ACM SIGMOD Rec. 14, 47–57 (1984).
Giannone, G. et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 99, 1303–1310 (2010).
Joensuu, M. et al. Visualizing endocytic recycling and trafficking in live neurons by subdiffractional tracking of internalized molecules. Nat. Protoc. 12, 2590–2622 (2017).
Joensuu, M. et al. Subdiffractional tracking of internalized molecules reveals heterogeneous motion states of synaptic vesicles. J. Cell Biol. 215, 277–292 (2016).
Willems, J. et al. ORANGE: a CRISPR–Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18, e3000665 (2020).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Erdmann, R. S. et al. Labeling strategies matter for super-resolution microscopy: a comparison between HaloTags and SNAP-tags. Cell Chem. Biol. 26, 584–592 e586 (2019).
de Beer, M. A. & Giepmans, B. N. G. Nanobody-based probes for subcellular protein identification and visualization. Front. Cell Neurosci. 14, 573278 (2020).
Sograte-Idrissi, S. et al. Circumvention of common labelling artefacts using secondary nanobodies. Nanoscale 12, 10226–10239 (2020).
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H. & Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012).
Prior, I. A., Muncke, C., Parton, R. G. & Hancock, J. F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003).
Nair, D. et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–13224 (2013).
Choquet, D. & Triller, A. The dynamic synapse. Neuron 80, 691–703 (2013).
Sieber, J. J. et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076 (2007).
Suzuki, K., Ritchie, K., Kajikawa, E., Fujiwara, T. & Kusumi, A. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88, 3659–3680 (2005).
Staus, D. P. et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535, 448–452 (2016).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Rasmussen, S. G. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011).
Irannejad, R. et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013).
Sungkaworn, T. et al. Single-molecule imaging reveals receptor–G protein interactions at cell surface hot spots. Nature 550, 543–547 (2017).
Maidorn, M., Olichon, A., Rizzoli, S. O. & Opazo, F. Nanobodies reveal an extra-synaptic population of SNAP-25 and Syntaxin 1A in hippocampal neurons. mAbs 11, 305–321 (2019).
Pardon, E. et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014).
Huang, R., Fang, P., Hao, Z. & Kay, B. K. Directed evolution of a highly specific FN3 monobody to the SH3 domain of human Lyn tyrosine kinase. PLoS ONE 11, e0145872 (2016).
Small, C. et al. SV2A controls the surface nanoclustering and endocytic recruitment of Syt1 during synaptic vesicle recycling. J. Neurochem. 168, 3188–3208 (2024).
Joensuu, M. et al. Presynaptic targeting of botulinum neurotoxin type A requires a tripartite PSG-Syt1-SV2 plasma membrane nanocluster for synaptic vesicle entry. EMBO J. 42, e112095 (2023).
Liau, W.-S. et al. Fear extinction is regulated by the activity of long noncoding RNAs at the synapse. Nat. Commun. 14, 7616 (2023).
Galli, V. et al. Uncoupling of dynamin polymerization and GTPase activity revealed by the conformation-specific nanobody dynab. eLife https://doi.org/10.7554/eLife.25197 (2017).
Jiang, A. et al. Dynamin1 long- and short-tail isoforms exploit distinct recruitment and spatial patterns to form endocytic nanoclusters. Nat. Commun. 15, 4060 (2024).
Cisse, I. I. et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667 (2013).
Lelek, M. et al. Single-molecule localization microscopy. Nat. Rev. Methods Primers https://doi.org/10.1038/s43586-021-00038-x (2021).
Khater, I. M., Nabi, I. R. & Hamarneh, G. A review of super-resolution single-molecule localization microscopy cluster analysis and quantification methods. Patterns 1, 100038 (2020).
Kubala, M. H., Kovtun, O., Alexandrov, K. & Collins, B. M. Structural and thermodynamic analysis of the GFP:GFP–nanobody complex. Protein Sci. 19, 2389–2401 (2010).
Kunz, P. et al. Exploiting sequence and stability information for directing nanobody stability engineering. Biochim. Biophys. Acta Gen. Subj. 1861, 2196–2205 (2017).
Dingus, J. G., Tang, J. C. Y., Amamoto, R., Wallick, G. K. & Cepko, C. L. A general approach for stabilizing nanobodies for intracellular expression. eLife https://doi.org/10.7554/eLife.68253 (2022).
Gross, G. G. et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985 (2013).
Tang, J. C. et al. Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. eLife https://doi.org/10.7554/eLife.15312 (2016).
Ariotti, N. et al. Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol. 16, e2005473 (2018).
Bademosi, A. T. et al. In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat. Commun. 8, 13660 (2017).
Gould, T. J. et al. Nanoscale imaging of molecular positions and anisotropies. Nat. Methods 5, 1027–1030 (2008).
Andronov, L., Lutz, Y., Vonesch, J. L. & Klaholz, B. P. SharpViSu: integrated analysis and segmentation of super-resolution microscopy data. Bioinformatics 32, 2239–2241 (2016).
Bayle, V. et al. Single-particle tracking photoactivated localization microscopy of membrane proteins in living plant tissues. Nat. Protoc. 16, 1600–1628 (2021).
De Zitter, E. et al. Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM. Nat. Methods 16, 707–710 (2019).
von Arx, M., Xhelilaj, K., Schulz, P., zur Oven-Krockhaus, S. & Gronnier, J. Photochromic reversion enables long-term tracking of single molecules in living plants. Preprint at bioRxiv https://doi.org/10.1101/2024.04.10.585335 (2024).
Tinevez, J. Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Zhang, M. et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9, 727–729 (2012).
Gotzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, 4403 (2019).
Durisic, N., Cuervo, L. L. & Lakadamyali, M. Quantitative super-resolution microscopy: pitfalls and strategies for image analysis. Curr. Opin. Chem. Biol. 20, 22–28 (2014).
van de Linde, S., Wolter, S., Heilemann, M. & Sauer, M. The effect of photoswitching kinetics and labeling densities on super-resolution fluorescence imaging. J. Biotechnol. 149, 260–266 (2010).
Bourgeois, D. Single molecule imaging simulations with advanced fluorophore photophysics. Commun. Biol. 6, 53 (2023).
Marsh, R. J. et al. Artifact-free high-density localization microscopy analysis. Nat. Methods 15, 689–692 (2018).
Waldchen, S., Lehmann, J., Klein, T., van de Linde, S. & Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 5, 15348 (2015).
Acknowledgements
This work is supported by a US National Institutes of Health R21 (grant no. RM2022000288) and a National Medical and Research Council (NHMRC) Fellowship (grant no. 1155794) awarded to F.A.M. T.P.W. is supported by an NHMRC Ideas Grant (grant no. 2010901) awarded to T.P.W and F.A.M. This work was supported by equipment funding from the Australian Research Council (ARC) (LE130100078), as well as funding from the University of Queensland Strategic Initiatives Fund grant no. DVCR22052A awarded to F.A.M. We thank the team at the QBI Advanced Microscopy Facility (University of Queensland) for their invaluable assistance. We sincerely thank all coauthors of the two preceding methods publications that form the basis of this Protocol’s manuscript.
Author information
Authors and Affiliations
Contributions
R.S.G., A.J., K.K. and S.F.L. performed the data processing and analysis. T.P.W. implemented the NASTIC workflow in Python. A.J.M. performed additional Python coding and GUI packaging. R.S.G. A.J.M., K.K., P.S and S.F.L. prepared the figures. R.S.G. and T.P.W. wrote the manuscript, with contributions from all authors. Preparation of the protocols for each section was as follows, tissue culture, transfection, plating and drift correction (R.S.G.); TIRF calibration, volume calculations and laser power assessment (K.K. and R.A.); SPT (P.S.); file conversion (A.J.M.) and NASTIC and segNASTIC (S.F.L. and T.P.W.). R.S.G., T.P.W. and F.A.M. designed the studies. The manuscript, figures and results were discussed with all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Dominique Bourgeois, Jip Wulffele, Akihiro Kusumi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Gormal et al. Proc. Natl Acad. Sci. USA 117 (48) 30476–30487 (2020): https://doi.org/10.1073/pnas.2007443117
Wallis et al. Nat. Commun. 14, 3353 (2023): https://doi.org/10.1038/s41467-023-38866-y
Joensuu et al. EMBO J. 42, e112095 (2023): https://doi.org/10.15252/embj.2022112095
Anmin et al. Nat. Commun. 15, 4060 (2024): https://doi.org/10.1038/s41467-024-47677-8
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1 and 2, Procedure and Manuals 1–6.
Supplementary Data 1 and 2 and Source Data Figs. 7 and 8
Example Wrangler summary tracking β2AR-YFP with GBP-mEos2. Example Wrangler summary tracking β2AR with Nb80-mEos2. Single particle tracking files (.trxyt) files as per Fig. 7 and Supplementary Data 1. Single particle tracking files (.trxyt) files as per Fig. 8 and Supplementary 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gormal, R.S., Wallis, T.P., McCann, A.J. et al. Nanoscale spatiotemporal cluster analysis of expressed and endogenous proteins. Nat Protoc 20, 3655–3694 (2025). https://doi.org/10.1038/s41596-025-01209-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01209-w