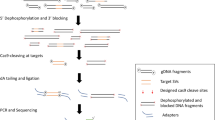
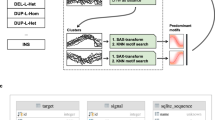
Abstract
Structural variants (SVs) contribute significantly to genomic diversity and disease predisposition as well as development in diverse species. However, their accurate characterization has remained a challenge because of their complexity and size. With the rise of third-generation sequencing technology, analytical strategies to map SVs have been revisited, and software such as NanoVar, a free and open-source package designed for efficient and reliable SV detection in long-read sequencing data, has facilitated their studies. NanoVar has been shown to work effectively in various published genomic studies, including research on genetic disorders, population genomics and genome analysis of non-model organisms. In this article, we describe in detail all the steps of the NanoVar protocol and its interplay with other platforms for SV calling in whole-genome long-read sequencing data such that researchers with minimal experience with command-line interfaces can easily carry out the protocol. It also provides exhaustive instructions for diverse study designs, including single-sample analyses, cohort studies and genome instability analyses. Finally, the protocol covers SV visualization, filtering and annotation details. Overall, users can identify and analyze SVs in a typical human dataset with a conventional computational setup in ~2–5 h after read mapping.
Key points
-
NanoVar is an optimized structural variant caller for long-read sequencing data and allows repeat element annotation of non-reference insertion variants.
-
This protocol provides researchers with a comprehensive structural variation detection guide from quality assessment of raw data to downstream analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The example dataset used in this protocol is from a published research article74 and is available at the Genome Sequence Archive for Human under accession number HRA002638. The protocol output directories are available on Zenodo at https://doi.org/10.5281/zenodo.1658352347.
Code availability
NanoVar and NanoINSight are publicly available at https://github.com/benoukraflab.
Change history
16 October 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41596-025-01297-8
References
Nesta, A. V., Tafur, D. & Beck, C. R. Hotspots of human mutation. Trends Genet. 37, 717–729 (2021).
Eichler, E. E. Genetic variation, comparative genomics, and the diagnosis of disease. N. Engl. J. Med. 381, 64–74 (2019).
Pang, A. W. et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11, R52 (2010).
Gilissen, C., Hoischen, A., Brunner, H. G. & Veltman, J. A. Unlocking Mendelian disease using exome sequencing. Genome Biol. 12, 228 (2011).
Logsdon, G. A., Vollger, M. R. & Eichler, E. E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 (2020).
Mantere, T., Kersten, S. & Hoischen, A. Long-read sequencing emerging in medical genetics. Front. Genet. 10, 426 (2019).
Merker, J. D. et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 20, 159–163 (2018).
Miao, H. et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 155, 32 (2018).
Loomis, E. W. et al. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 23, 121–128 (2013).
Schüle, B. et al. Parkinson’s disease associated with pure ATXN10 repeat expansion. NPJ Parkinson’s Dis. 3, 27 (2017).
Höijer, I. et al. Detailed analysis of HTT repeat elements in human blood using targeted amplification-free long-read sequencing. Hum. Mutat. 39, 1262–1272 (2018).
Cumming, S. A. et al. De novo repeat interruptions are associated with reduced somatic instability and mild or absent clinical features in myotonic dystrophy type 1. Eur. J. Hum. Genet. 26, 1635–1647 (2018).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Hoyt, S. J. et al. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science 376, eabk3112 (2022).
Ayarpadikannan, S. & Kim, H.-S. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 12, 98–104 (2014).
Hancks, D. C. & Kazazian, H. H. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 (2016).
Gardner, E. J. et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 27, 1916–1929 (2017).
Thung, D. T. et al. Mobster: accurate detection of mobile element insertions in next generation sequencing data. Genome Biol. 15, 488 (2014).
Tubio, J. M. C. et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345, 1251343 (2014).
Torene, R. I. et al. Mobile element insertion detection in 89,874 clinical exomes. Genet. Med. 22, 974–978 (2020).
Shiraishi, Y. et al. Precise characterization of somatic complex structural variations from tumor/control paired long-read sequencing data with nanomonsv. Nucleic Acids Res. 51, e74 (2023).
Lei, Y. et al. Overview of structural variation calling: simulation, identification, and visualization. Comput. Biol. Med. 145, 105534 (2022).
De Coster, W. et al. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 29, 1178–1187 (2019).
Yang, L. A practical guide for structural variation detection in human genome. Curr. Protoc. Hum. Genet. 107, e103 (2020).
Tham, C. Y. et al. NanoVar: accurate characterization of patients’ genomic structural variants using low-depth nanopore sequencing. Genome Biol. 21, 56 (2020).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Smit, AFA, Hubley, R. & Green, P. RepeatMasker Open-4.0 (2013–2015).
Cretu Stancu, M. et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 8, 1326 (2017).
Gong, L. et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods 15, 455–460 (2018).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single molecule sequencing. Nat. Methods 15, 461–468 (2018).
Smolka, M. et al. Detection of mosaic and population-level structural variants with Sniffles2. Nat. Biotechnol. 42, 1571–1580 (2024).
Jiang, T. et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 21, 189 (2020).
Jiang, T. et al. cuteFC: regenotyping structural variants through an accurate and efficient force-calling method. Genome Biol. 26, 166 (2025).
Jiang, T. et al. Long-read sequencing settings for efficient structural variation detection based on comprehensive evaluation. BMC Bioinforma. 22, 552 (2021).
Heller, D. & Vingron, M. SVIM: structural variant identification using mapped long reads. Bioinformatics 35, 2907–2915 (2019).
Tham, C. Y. & Benoukraf, T. Correspondence on NanoVar’s performance outlined by Jiang T. et al. in “Long-read sequencing settings for efficient structural variation detection based on comprehensive evaluation”. BMC Bioinformatics 24, 350 (2023).
Dierckxsens, N., Li, T., Vermeesch, J. R. & Xie, Z. A benchmark of structural variation detection by long reads through a realistic simulated model. Genome Biol. 22, 342 (2021).
Wu, Z. et al. Structural variants in the Chinese population and their impact on phenotypes, diseases and population adaptation. Nat. Commun. 12, 6501 (2021).
Liu, Y. H., Luo, C., Golding, S. G., Ioffe, J. B. & Zhou, X. M. Tradeoffs in alignment and assembly-based methods for structural variant detection with long-read sequencing data. Nat. Commun. 15, 2447 (2024).
Liu, Y. et al. Comparison of structural variants detected by PacBio-CLR and ONT sequencing in pear. BMC Genomics 23, 830 (2022).
Fiol, A., Jurado-Ruiz, F., López-Girona, E. & Aranzana, M. J. An efficient CRISPR-Cas9 enrichment sequencing strategy for characterizing complex and highly duplicated genomic regions. A case study in the Prunus salicina LG3-MYB10 genes cluster. Plant Methods 18, 105 (2022).
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018).
De Coster, W. & Rademakers, R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 39, btad311 (2023).
Asmaa, S., Tham, C. Y., Dyer, M. & Benoukraf, T. Dataset for ‘NanoVar: a Comprehensive Workflow for Structural Variant Detection to uncover the Genome’s Hidden Patterns’. Zenodo https://zenodo.org/records/16583523 (2025).
Kiełbasa, S. M., Wan, R., Sato, K., Horton, P. & Frith, M. C. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011).
Sović, I. et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 7, 11307 (2016).
Zhou, A., Lin, T. & Xing, J. Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol. 20, 237 (2019).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Jeffares, D. C. et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 8, 14061 (2017).
English, A. C., Menon, V. K., Gibbs, R. A., Metcalf, G. A. & Sedlazeck, F. J. Truvari: refined structural variant comparison preserves allelic diversity. Genome Biol. 23, 271 (2022).
Kirsche, M. et al. Jasmine and Iris: population-scale structural variant comparison and analysis. Nat. Methods 20, 408–417 (2023).
Zheng, Z. et al. A sequence-aware merger of genomic structural variations at population scale. Nat. Commun. 15, 960 (2024).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Yates, A. et al. The Ensembl REST API: Ensembl data for any language. Bioinformatics 31, 143–145 (2015).
Geoffroy, V. et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 34, 3572–3574 (2018).
Cunningham, F., Moore, B., Ruiz-Schultz, N., Ritchie, G. R. & Eilbeck, K. Improving the Sequence Ontology terminology for genomic variant annotation. J. Biomed. Semant. 6, 32 (2015).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Zwaig, M. et al. Linked-read based analysis of the medulloblastoma genome. Front. Oncol. 13, 1221611 (2023).
Klever, M.-K. et al. AML with complex karyotype: extreme genomic complexity revealed by combined long-read sequencing and Hi-C technology. Blood Adv. 7, 6520–6531 (2023).
Greer, S. U. et al. Implementation of Nanopore sequencing as a pragmatic workflow for copy number variant confirmation in the clinic. J. Transl. Med. 21, 378 (2023).
Gladysheva-Azgari, M. et al. A de novo genome assembly of cultivated Prunus persica cv. ‘Sovetskiy’. PLoS ONE 17, e0269284 (2022).
Ji, C.-M., Feng, X.-Y., Huang, Y.-W. & Chen, R.-A. The applications of nanopore sequencing technology in animal and human virus research. Viruses 16, 798 (2024).
Elrick, H. et al. SAVANA: reliable analysis of somatic structural variants and copy number aberrations using long-read sequencing. Nat. Methods 22, 1436–1446 (2025).
Keskus, A. G. et al. Severus detects somatic structural variation and complex rearrangements in cancer genomes using long-read sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02618-8 (2025).
Liu, L. et al. Performance of somatic structural variant calling in lung cancer using Oxford Nanopore sequencing technology. BMC Genomics 25, 898 (2024).
Cameron, D. L., Di Stefano, L. & Papenfuss, A. T. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 10, 3240 (2019).
Kosugi, S. et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 20, 117 (2019).
Alioto, T. S. et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat. Commun. 6, 10001 (2015).
Ewing, A. D. et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods 12, 623–630 (2015).
Cuenca-Guardiola, J. et al. Detection and annotation of transposable element insertions and deletions on the human genome using nanopore sequencing. iScience 26, 108214 (2023).
Xu, L. et al. Long-read sequencing identifies novel structural variations in colorectal cancer. PLoS Genet. 19, e1010514 (2023).
Liu, Z., Xie, Z. & Li, M. Comprehensive and deep evaluation of structural variation detection pipelines with third-generation sequencing data. Genome Biol. 25, 188 (2024).
Quan, C., Lu, H., Lu, Y. & Zhou, G. Population-scale genotyping of structural variation in the era of long-read sequencing. Comput. Struct. Biotechnol. J. 20, 2639–2647 (2022).
Aganezov, S. et al. A complete reference genome improves analysis of human genetic variation. Science 376, eabl3533 (2022).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Zhao, P., Li, L., Jiang, X. & Li, Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J. Hematol. Oncol. 12, 54 (2019).
Cornish, A. J. et al. The genomic landscape of 2,023 colorectal cancers. Nature 633, 127–136 (2024).
Acknowledgements
The authors thank H. Alloway for her proof reading as well as the Centre for Analytics, Informatics and Research (CAIR) at Memorial University and the Digital Research Alliance of Canada (the Alliance) in partnership with ACENET, for their support in providing high-performance computing resources that facilitated the development and testing of our protocol.
Author information
Authors and Affiliations
Contributions
A.S. and C.Y.T. conceived and designed the protocol. M.D. tested the protocol and reported issues. A.S. and T.B. wrote the manuscript. T.B. supervised the design of the protocol.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Andrew Beggs and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key reference
Tham, C.Y. et al. Genome Biol. 21, 56 (2020): https://doi.org/10.1186/s13059-020-01968-7
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Samy, A., Tham, C.Y., Dyer, M. et al. NanoVar: a comprehensive workflow for structural variant detection to uncover the genome’s hidden patterns. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01270-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01270-5