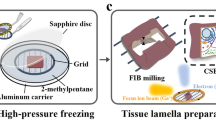
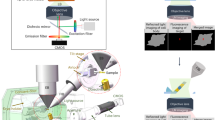
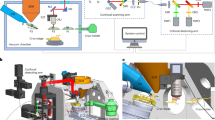
Abstract
Cryogenic-electron tomography (cryo-ET) permits the in situ visualization of biological macromolecules at the molecular level. Owing to the variable thickness of cells, tissues and organisms, frozen specimens may need to be thinned by cryo-focused ion beam (FIB) milling to produce thin (<500 nm) cryo-lamellae suitable for cryo-ET. Locating regions of interest remains a challenge because untargeted milling can lead to inadvertent ablation and removal of regions of interest. Correlative light and electron microscopy, combined with cryo-FIB milling, can guide the identification of labeled targets in the cellular milieu. Multiple transfers between cryo-imaging instruments, cumbersome correlation algorithms, limited accuracy and low throughput have hindered the routine adoption of cryo-FIB milling within a multimodal correlative workflow for in situ structural biology. Here we present a workflow for 3D correlative cryo-fluorescence light microscopy-FIB-ET that streamlines fluorescence light microscopy-guided FIB milling, improving throughput while preserving both structural and contextual information. The complete integration of hardware and software described here minimizes sample contamination from cross-platform exchanges and greatly enhances the efficiency of 3D targeting in cryo-milling. We then describe procedures for implementing montage parallel array cryo-ET (MPACT), which can be easily adapted to any modern life-science transmission electron microscope. MPACT supports high-throughput cryo-ET acquisitions (10 tilt series in 1.5 h) for structure determination and comprehensive contextual understanding of macromolecules within their native surroundings. A complete session from sample preparation to MPACT data processing takes 5−7 d for an individual experienced in both cryo-EM and cryo-FIB milling.
Key points
-
Coupling 3D correlative focused ion beam milling with montage cryo-electron tomography enables the precise localization of regions of interest deep within cells and the acquisition of larger fields of view that preserve high-resolution structural details.
-
Single-pair region of interest-marker registration, combined with integrated software and hardware, enhances correlative milling workflows. Montage parallel array cryo-electron tomography produces montages and individual tile tomograms, thereby eliminating possible mechanical adjustments to the transmission electron microscope.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
Data availability
All relevant data are available from the corresponding author upon request. The iFLM image stacks and corresponding SEM and FIB images are associated with the on-the-fly 3D correlation procedure. While it is challenging to share, a demonstration could be performed to help the user upon reasonable request. A set of raw frames for a representative 3 × 3 MPACT montage tilt series is provided as a tutorial dataset and available to download at https://github.com/wright-cemrc-projects/cryoet-montage/tree/main/Tutorial/ for demonstration of MPACT preprocessing, stitching and individual tile tilt series generation. Subtomogram averages of respiratory syncytial virus fusion pair particles using MPACT individual tile tilt series have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-40308 and EMD-40307.
Code availability
CorRelator and TomoGrapher are open-source software, and the source code can be downloaded via https://github.com/wright-cemrc-projects. The MPACT processing scripts can be downloaded via https://github.com/wright-cemrc-projects/cryoet-montage/tree/main/Bashscripts.
References
Hong, Y., Song, Y., Zhang, Z. & Li, S. Cryo-electron tomography: the resolution revolution and a surge of in situ virological discoveries. Annu. Rev. Biophys. 52, 339–360 (2023).
Turk, M. & Baumeister, W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 594, 3243–3261 (2020).
Al-Amoudi, A. et al. Cryo-electron microscopy of vitreous sections. EMBO J. 23, 3583–3588 (2004).
Berger, C. et al. Cryo-electron tomography on focused ion beam lamellae transforms structural cell biology. Nat. Methods 20, 499–511 (2023).
Rigort, A. et al. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 172, 169–179 (2010).
Hampton, C. M. et al. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat. Protoc. 12, 150–167 (2017).
Yang, J. E., Larson, M. R., Sibert, B. S., Shrum, S. & Wright, E. R. CorRelator: interactive software for real-time high precision cryo-correlative light and electron microscopy. J. Struct. Biol. 213, 107709 (2021).
Smeets, M. et al. Integrated cryo-correlative microscopy for targeted structural investigation in situ. Microsc. Today 29, 20–25 (2021).
Arnold, J. et al. Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 110, 860–869 (2016).
Hall, A. S., Lavery, L. L. & Doux, P. Effective multimodal multiscale analytical and imaging correlation. IEEE Sens. Lett. 3, 1–4 (2019).
Moser, F. et al. Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. Proc. Natl Acad. Sci. USA 116, 4804–4809 (2019).
Schorb, M. et al. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. J. Struct. Biol. 197, 83–93 (2017).
Chang, Y.-W. et al. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 11, 737–739 (2014).
Fukuda, Y. et al. Coordinate transformation based cryo-correlative methods for electron tomography and focused ion beam milling. Ultramicroscopy 143, 15–23 (2014).
Kelley, K. et al. Waffle Method: a general and flexible approach for improving throughput in FIB-milling. Nat. Commun. 13, 1857 (2022).
Wolff, G. et al. Mind the gap: micro-expansion joints drastically decrease the bending of FIB-milled cryo-lamellae. J. Struct. Biol. 208, 107389 (2019).
Booy, F. P. & Pawley, J. B. Cryo-crinkling: what happens to carbon films on copper grids at low temperature. Ultramicroscopy 48, 273–280 (1993).
Li, W. et al. Integrated multimodality microscope for accurate and efficient target-guided cryo-lamellae preparation. Nat. Methods 20, 268–275 (2023).
Li, S. et al. ELI trifocal microscope: a precise system to prepare target cryo-lamellae for in situ cryo-ET study. Nat. Methods 20, 276–283 (2023).
Gorelick, S. et al. PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. eLife 8, e45919 (2019).
Tacke, S. et al. A streamlined workflow for automated cryo focused ion beam milling. J. Struct. Biol. 213, 107743 (2021).
Klumpe, S. et al. A modular platform for automated cryo-FIB workflows. eLife 10 (2021).
Zachs, T. et al. Fully automated, sequential focused ion beam milling for cryo-electron tomography. eLife 9 (2020).
Hylton, R. K. & Swulius, M. T. Challenges and triumphs in cryo-electron tomography. iScience 24, 102959 (2021).
Schur, F. K. M. Toward high-resolution in situ structural biology with cryo-electron tomography and subtomogram averaging. Curr. Opin. Struct. Biol. 58, 1–9 (2019).
Yang, J. E. et al. Correlative montage parallel array cryo-tomography for in situ structural cell biology. Nat. Methods 20, 1537–1543 (2023).
Huang, Y.-W., Cambre, M. & Lee, H.-J. The toxicity of nanoparticles depends on multiple molecular and physicochemical mechanisms. Int. J. Mol. Sci. 18, 2702 (2017).
Scher, N., Rechav, K., Paul-Gilloteaux, P. & Avinoam, O. In situ fiducial markers for 3D correlative cryo-fluorescence and FIB-SEM imaging. iScience 24, 102714 (2021).
Yang, J. E., Mitchell, J. M., Bingman, C. A., Mosher, D. F. & Wright, E. R. In situ crystalline structure of the human eosinophil major basic protein-1. Preprint at bioRxiv https://doi.org/10.1101/2024.10.09.617336 (2024).
Kim, J. Y. et al. Handling difficult cryo-ET samples: a study with primary neurons from Drosophila melanogaster. Microsc. Microanal. 29, 2127–2148 (2023).
Sibert, B. S., Kim, J. Y., Yang, J. E. & Wright, E. R. Micropatterning transmission electron microscopy grids to direct cell positioning within whole-cell cryo-electron tomography workflows. J. Vis. Exp. 175, e62992 (2021).
Sibert, B. S. et al. Assembly of respiratory syncytial virus matrix protein lattice and its coordination with fusion glycoprotein trimers. Nat. Commun. 15, 5923 (2024).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Mastronarde, D. Tomographic reconstruction with the IMOD software package. Microsc. Microanal. 12, 178–179 (2006).
Garza-Lopez, E. et al. Protocols for generating surfaces and measuring 3D organelle morphology using Amira. Cells 11, 65 (2022).
Stalling, D., Westerhoff, M. & Hege, H.-C. in The Visualization Handbook (Academic Press, 2005).
Chen, M. et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14, 983–985 (2017).
Heebner, J. E. et al. Deep learning-based segmentation of cryo-electron tomograms. J. Vis. Exp. 189, e64435 (2022).
Wan, W. & Briggs, J. A. Cryo-electron tomography and subtomogram averaging. Methods Enzymol. 579, 329–367 (2016).
Castaño-Díez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012).
Nicastro, D. et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006).
Bharat, T. A. M. & Scheres, S. H. W. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat. Protoc. 11, 2054–2065 (2016).
Wagner, F. R. et al. Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat. Protoc. 15, 2041–2070 (2020).
Boltje, D. B. et al. A cryogenic, coincident fluorescence, electron, and ion beam microscope. eLife 11, e82891 (2022).
Schiøtz, O. H. et al. Serial Lift-Out: sampling the molecular anatomy of whole organisms. Nat. Methods 21, 1684–1692 (2023).
Paul-Gilloteaux, P. et al. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat. Methods 14, 102–103 (2017).
Schorb, M. & Briggs, J. A. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 143, 24–32 (2014).
Kukulski, W. et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 192, 111–119 (2011).
Peck, A. et al. Montage electron tomography of vitrified specimens. J. Struct. Biol. 214, 107860 (2022).
Elferich, J., Schiroli, G., Scadden, D. T. & Grigorieff, N. Defocus Corrected Large Area Cryo-EM (DeCo-LACE) for label-free detection of molecules across entire cell sections. eLife 11, e80980 (2022).
Chua, E. Y. D. et al. Square beams for optimal tiling in transmission electron microscopy. Nat. Methods 21, 562–565 (2024).
Brown, H. G., Smith, D., Wardle, B. C. & Hanssen, E. Square condenser apertures for square cameras in low-dose transmission electron microscopy. Nat. Methods 21, 566–568 (2024).
Eisenstein, F. et al. Parallel cryo electron tomography on in situ lamellae. Nat. Methods 20, 131–138 (2023).
Bouvette, J. et al. Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography. Nat. Commun. 12, 1957 (2021).
Eisenstein, F., Danev, R. & Pilhofer, M. Improved applicability and robustness of fast cryo-electron tomography data acquisition. J. Struct. Biol. 208, 107–114 (2019).
Chreifi, G., Chen, S., Metskas, L. A., Kaplan, M. & Jensen, G. J. Rapid tilt-series acquisition for electron cryotomography. J. Struct. Biol. 205, 163–169 (2019).
Sexton, D. L., Burgold, S., Schertel, A. & Tocheva, E. I. Super-resolution confocal cryo-CLEM with cryo-FIB milling for in situ imaging of Deinococcus radiodurans. Curr. Res. Struct. Biol. 4, 1–9 (2022).
Weber, N., Hinks, B., Jensen, J., Lidahl, T. & Mendonça, L. Sample preparation for in situ cryotomography of mammalian cells. J. Vis. Exp. 202, e65697 (2023).
Thompson, R. F., Walker, M., Siebert, C. A., Muench, S. P. & Ranson, N. A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 100, 3–15 (2016).
de Beer, M. et al. Precise targeting for 3D cryo-correlative light and electron microscopy volume imaging of tissues using a FinderTOP. Commun. Biol. 6, 510 (2023).
Toro-Nahuelpan, M. et al. Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Nat. Methods 17, 50–54 (2020).
Bäuerlein, F. J. B. et al. Cryo-electron tomography of large biological specimens vitrified by plunge freezing. Preprint at bioRxiv https://doi.org/10.1101/2021.04.14.437159 (2023).
Hands-Portman, I. & Bakker, S. E. Customising the plunge-freezing workflow for challenging conditions. Faraday Discuss. 240, 44–54 (2022).
Tuijtel, M. W., Koster, A. J., Jakobs, S., Faas, F. G. A. & Sharp, T. H. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep. 9, 1369 (2019).
Metskas, L. A. & Briggs, J. A. G. Fluorescence-based detection of membrane fusion state on a cryo-EM grid using correlated cryo-fluorescence and cryo-electron microscopy. Microsc. Microanal. 25, 942–949 (2019).
Karuppasamy, M., Karimi Nejadasl, F., Vulovic, M., Koster, A. J. & Ravelli, R. B. Radiation damage in single-particle cryo-electron microscopy: effects of dose and dose rate. J. Synchrotron Radiat. 18, 398–412 (2011).
Basanta, B., Hirschi, M. M., Grotjahn, D. A. & Lander, G. C. A case for glycerol as an acceptable additive for single-particle cryoEM samples. Acta Crystallogr. D. 78, 124–135 (2022).
Melo, R. C. N. & Weller, P. F. Contemporary understanding of the secretory granules in human eosinophils. J. Leukoc. Biol. 104, 85–93 (2018).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Chen, M. et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168 (2019).
Tegunov, D., Xue, L., Dienemann, C., Cramer, P. & Mahamid, J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat. Methods 18, 186–193 (2021).
Dimchev, G., Amiri, B., Fäßler, F., Falcke, M. & Schur, F. K. Computational toolbox for ultrastructural quantitative analysis of filament networks in cryo-ET data. J. Struct. Biol. 213, 107808 (2021).
Lam, V. & Villa, E. Practical approaches for cryo-FIB milling and applications for cellular cryo-electron tomography. Methods Mol. Biol. 2215, 49–82 (2021).
Rigort, A. et al. Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J. Struct. Biol. 177, 135–144 (2012).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Schaffer, M. et al. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197, 73–82 (2017).
Langford, R. M. Focused ion beam nanofabrication: a comparison with conventional processing techniques. J. Nanosci. Nanotechnol. 6, 661–668 (2006).
Acknowledgements
We are grateful for the TEM and SEM instrumentation support of M. Woods and T. Coomes from Thermo Fisher Scientific. We are thankful for the computational resources supplied through the SBGrid76. This work was supported in part by the University of Wisconsin, Madison, the Department of Biochemistry at the University of Wisconsin, Madison, the Morgridge Institute for Research, and public health service grant nos. R01 GM114561, R01 GM132068, RF1 NS110436 and U24 GM139168 to E.R.W., P01 HL088594 to N. Jarjour, and R01 AI125390 to D.F.M. and J. Coon from the National Institutes of Health. J.M.M. was supported by the grant T32 HL07899 from the National Heart, Lung, and Blood Institute, National Institutes of Health. This work was supported in part by the US Department of Energy, Office of Science, Office of Biological and Environmental Research under award number DE-SC0018409. We are grateful for the use of facilities and instrumentation at the Cryo-EM Research Center and the Midwest Center for Cryo-ET in the Department of Biochemistry at the University of Wisconsin, Madison.
Author information
Authors and Affiliations
Contributions
E.R.W., J.M., J.E.Y., V.V., T.F. and A.R. conceived the study. J.E.Y. and E.R.W. designed the study. J.E.Y., V.V., B.S.S., J.M.M., T.F., A.S.H., A.R., M.R.L., D.F.M., J.M. and E.R.W. prepared the samples, performed the experiments and processed the data. J.E.Y. and E.R.W. wrote the manuscript with contributions from all authors. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
V.V., T.F., A.H., A.R. and J.M. are employees of Thermo Fisher Scientific. Other authors have no competing interests or other interests that might be perceived to influence the interpretation of the article.
Peer review
Peer review information
Nature Protocols thanks Johannes Groen, Michael Martynowycz and Anna Steyer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Hampton, C. M. et al. Nat. Protoc. 12, 150–167 (2017): https://doi.org/10.1038/nprot.2016.168
Yang, J. E. et al. J. Struct. Biol. 213, 107709 (2021): https://doi.org/10.1016/j.jsb.2021.107709
Sibert, B. S. et al. J. Vis. Exp. 175, 10.3791/62992 (2021): https://doi.org/10.3791/62992
Yang, J. E. et al. Nat. Methods 20, 1537 (2023): https://doi.org/10.1038/s41592-023-01999-5
Kim, J. Y. et al. Microsc. Microanal. 29, 2127 (2023): https://doi.org/10.1093/micmic/ozad125
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J.E., Vrbovská, V., Mitchell, J.M. et al. Integrated fluorescence light microscopy-guided cryo-focused ion beam-milling for in situ montage cryo-ET. Nat Protoc (2026). https://doi.org/10.1038/s41596-025-01284-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01284-z