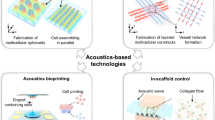
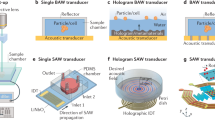
Abstract
The isolation of small extracellular vesicles (sEVs), viruses and other nanoscale lipid particles from biofluids offers actionable possibilities for advancing disease diagnosis, drug delivery, regenerative medicine, personalized medicine and immunotherapy. Several methods are available to isolate sEVs from biofluids and acoustic techniques provide distinct advantages. Challenges constraining its wider application encompass the absence of adequate procedures for fabrication, implementation and performance validation. These issues impede the development of protocols applicable to nanoscale bioparticles experiencing acoustic isolation effects. Here we present a detailed protocol for acoustic separation of nanoscale bioparticles from biofluids, including plasma and saliva, achieving both high purity and throughput suitable for routine application. This protocol offers a comprehensive, step-by-step guide for the design and fabrication of the acoustic separation device, the establishment of the experimental setup and the isolation of bioparticles. To ensure reliability, rigor and reproducibility, we delineate essential procedures, including acoustic field optimization, channel fabrication and biofluid preparation, subsequently validating the protocol and its performance across different operators. Our protocol further encompasses procedures for data collection and analysis, which are essential for characterizing viruses and sEVs, as well as for evaluating their quality and integrity. This protocol enables researchers to perform high-quality isolation of nanoscale bioparticles, providing access to reliable acoustic separation techniques. Standardizing this technique will pave the way for discoveries in virology and intercellular communication research, with applications in medicine, biology, and materials science.
Key points
-
This protocol provides a guide for designing and fabricating the acoustic separation device, including channel fabrication, the experimental setup and acoustic field optimization, and the biofluid preparation and isolation of nano-sized biological particles.
-
Alternative approaches for biological nanoparticle separation include differential ultracentrifugation; however, this has a low yield and is time consuming and labor intensive. Another alternative is size exclusion chromatography, which has drawbacks such as sample dilution and limited resolution below 70 nm.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the article. The sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE235349. Further information is available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Witwer, K. W. & Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 8, 1648167 (2019).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
El Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. A. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Dai, J. et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 5, 145 (2020).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967––1976 (2003).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C. & Garry, R. F. The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020).
Gire, S. K. et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 (2014).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Su, J. et al. Cell–cell communication: new insights and clinical implications. Signal Transduct. Target. Ther. 9, 196 (2024).
Jin, S. et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Théry, C., Ostrowski, M. & Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009).
Gonzales, P. A. et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 20 (2009).
Menachery, V. D. et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513 (2015).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Fang, T. et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 9, 191 (2018).
Al-Nedawi, K. et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 10, 619–624 (2008).
Liu, M., Hu, S., Yan, N., Popowski, K. D. & Cheng, K. Inhalable extracellular vesicle delivery of IL-12 mRNA to treat lung cancer and promote systemic immunity. Nat. Nanotechnol. 19, 565–575 (2024).
Wang, Z. et al. Inhalation of ACE2-expressing lung exosomes provides prophylactic protection against SARS-CoV-2. Nat. Commun. 15, 2236 (2024).
Margolis, L. B. & Sadovsky, Y. When extracellular vesicles go viral: a bird’s eye view. Pathog. Immun. 10, 140–158 (2024).
Momen-Heravi, F. in Extracellular Vesicles: Methods and Protocols (eds Kuo, W. P. & Jia, S.) 25–32 (Springer, 2017).
Jia, Y. et al. Small extracellular vesicles isolation and separation: current techniques, pending questions and clinical applications. Theranostics 12, 6548–6575 (2022).
Sugita, Y., Noda, T., Sagara, H. & Kawaoka, Y. Ultracentrifugation deforms unfixed influenza A virions. J. Gen. Virol. 92, 2485–2493 (2011).
Gias, E., Nielsen, S. U., Morgan, L. A. F. & Toms, G. L. Purification of human respiratory syncytial virus by ultracentrifugation in iodixanol density gradient. J. Virol. Methods 147, 328–332 (2008).
Bergqvist, M., Lässer, C., Crescitelli, R., Park, K.-S. & Lötvall, J. A non-centrifugation method to concentrate and purify extracellular vesicles using superabsorbent polymer followed by size exclusion chromatography. J. Extracell. Vesicles 14, e70037 (2025).
Sidhom, K., Obi, P. O. & Saleem, A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int. J. Mol. Sci. 21, 6466 (2020).
Stott, S. L. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA 107, 18392–18397 (2010).
Zhang, P. et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 3, 438–451 (2019).
Rabe, D. C. et al. Ultrasensitive detection of intact SARS-CoV-2 particles in complex biofluids using microfluidic affinity capture. Sci. Adv. 11, eadh1167 (2025).
Sunkara, V. et al. Fully automated, label-free isolation of extracellular vesicles from whole blood for cancer diagnosis and monitoring. Theranostics 9, 1851–1863 (2019).
Hu, J. & Gao, D. Recent advances in aptamer-based microfluidic biosensors for the isolation, signal amplification and detection of exosomes. Sensors 25, 848 (2025).
Im, H. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 32, 490–495 (2014).
Reátegui, E. et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 9, 175 (2018).
Huang, L. R., Cox, E. C., Austin, R. H. & Sturm, J. C. Continuous particle separation through deterministic lateral displacement. Science 304, 987–990 (2004).
Wunsch, B. H. et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 11, 936–940 (2016).
Shi, J., Huang, H., Stratton, Z., Huang, Y. & Huang, T. J. Continuous particle separation in a microfluidic channelvia standing surface acoustic waves (SSAW). Lab Chip 9, 3354–3359 (2009).
Ding, X. et al. Cell separation using tilted-angle standing surface acoustic waves. Proc. Natl Acad. Sci. USA 111, 12992–12997 (2014).
Li, P. et al. Acoustic separation of circulating tumor cells. Proc. Natl Acad. Sci. USA 112, 4970–4975 (2015).
Wu, M. et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl Acad. Sci. USA 114, 10584–10589 (2017).
Wu, M. et al. Acoustofluidic separation of cells and particles. Microsyst. Nanoeng. 5, 32 (2019).
Wang, Z. et al. Acoustofluidic salivary exosome isolation: a liquid biopsy compatible approach for human papillomavirus–associated oropharyngeal cancer detection. J. Mol. Diagn. 22, 50–59 (2020).
Gu, Y. et al. Acoustofluidic centrifuge for nanoparticle enrichment and separation. Sci. Adv. 7, eabc0467 (2021).
Xia, J. et al. Acoustofluidic virus isolation via bessel beam excitation separation technology. ACS Nano 18, 22596–22607 (2024).
Naquin, T. D. et al. Acoustic separation and concentration of exosomes for nucleotide detection: ASCENDx. Sci. Adv. 10, eadm8597 (2024).
Wang, Z. et al. Acoustofluidic separation enables early diagnosis of traumatic brain injury based on circulating exosomes. Microsyst. Nanoeng. 7, 20 (2021).
Choi, W. et al. CDK1-loaded extracellular vesicles promote cell cycle to reverse impaired wound healing in diabetic obese mice. Mol. Ther. 33, 1118–1133 (2025).
Welsh, J. A. et al. MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles 9, 1713526 (2020).
Welsh, J. A. et al. A compendium of single extracellular vesicle flow cytometry. J. Extracell. Vesicles 12, e12299 (2023).
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Rozowsky, J. et al. exceRpt: a comprehensive analytic platform for extracellular RNA profiling. Cell Syst. 8, 352–357.e353 (2019).
Hinestrosa, J. P. et al. Early-stage multi-cancer detection using an extracellular vesicle protein-based blood test. Commun. Med. 2, 29 (2022).
Sigdel, S., Swenson, S. & Wang, J. Extracellular vesicles in neurodegenerative diseases: an update. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241713161 (2023).
Zarà, M. et al. Circulating small extracellular vesicles reflect the severity of myocardial damage in STEMI patients. Biomolecules 13, 1470 (2023).
Cruz, C. G., Sodawalla, H. M., Mohanakumar, T. & Bansal, S. Extracellular vesicles as biomarkers in infectious diseases. Biology 14, 182 (2025).
Sun, M. et al. Extracellular vesicles: a new star for gene drug delivery. Int. J. Nanomed. 19, 2241–2264 (2024).
Lundstrom, K. Viral vectors in gene therapy. Diseases https://doi.org/10.3390/diseases6020042 (2018).
Berumen Sánchez, G., Bunn, K. E., Pua, H. H. & Rafat, M. Extracellular vesicles: mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 19, 104 (2021).
Xu, J. et al. Human perivascular stem cell-derived extracellular vesicles mediate bone repair. eLife 8, e48191 (2019).
Walker, L. M. & Burton, D. R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 18, 297–308 (2018).
Raab-Traub, N. & Dittmer, D. P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 15, 559–572 (2017).
Li, P., Kaslan, M., Lee, S. H., Yao, J. & Gao, Z. Progress in exosome isolation techniques. Theranostics 7, 789–804 (2017).
Kowal, J. et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl Acad. Sci. USA 113, E968–E977 (2016).
Yang, Y. et al. Extracellular vesicles isolated by size-exclusion chromatography present suitability for RNomics analysis in plasma. J. Transl. Med. 19, 104 (2021).
Rezeli, M. et al. Comparative proteomic analysis of extracellular vesicles isolated by acoustic trapping or differential centrifugation. Anal. Chem. 88, 8577–8586 (2016).
Alexandre, L. et al. Effect of sample preprocessing and size-based extraction methods on the physical and molecular profiles of extracellular vesicles. ACS Sens. 9, 1239–1251 (2024).
Chen, J. et al. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. https://doi.org/10.3389/fbioe.2021.811971 (2022).
Wullenweber, M. S., Kottmeier, J., Kampen, I., Dietzel, A. & Kwade, A. Simulative investigation of different DLD microsystem designs with increased Reynolds numbers using a two-way coupled IBM-CFD/6-DOF approach. Processes 10, 403 (2022).
Zhang, J. et al. Fundamentals and applications of inertial microfluidics: a review. Lab Chip 16, 10–34 (2016).
Nguyen, K. T. et al. Integrated techniques for extracellular particle separation and single-particle multiparametric characterization to track cancer biomarkers from tissue to biofluids. Preprint at bioRxiv https://doi.org/10.1101/2025.01.09.632270 (2025).
Cai, S. et al. Artificial intelligence velocimetry and microaneurysm-on-a-chip for three-dimensional analysis of blood flow in physiology and disease. Proc. Natl Acad. Sci. USA 118, e2100697118 (2021).
Yeager, M., Wilson-Kubalek, E. M., Weiner, S. G., Brown, P. O. & Rein, A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: Implications for retroviral assembly mechanisms. Proc. Natl Acad. Sci. USA 95, 7299–7304 (1998).
Wu, M. et al. Separating extracellular vesicles and lipoproteins via acoustofluidics. Lab Chip 19, 1174–1182 (2019).
Xie, Y. et al. Acoustic cell separation based on density and mechanical properties. J. Biomech. Eng. https://doi.org/10.1115/1.4046180 (2020).
Friend, J. & Yeo, L. Y. Microscale acoustofluidics: microfluidics driven via acoustics and ultrasonics. Rev. Mod. Phys. 83, 647–704 (2011).
Del Campo Fonseca, A. et al. Ultrasound trapping and navigation of microrobots in the mouse brain vasculature. Nat. Commun. 14, 5889 (2023).
Collins, D. J., O’Rorke, R., Neild, A., Han, J. & Ai, Y. Acoustic fields and microfluidic patterning around embedded micro-structures subject to surface acoustic waves. Soft Matter 15, 8691–8705 (2019).
Cai, H. et al. Brain organoid reservoir computing for artificial intelligence. Nat. Electron. 6, 1032–1039 (2023).
Drinkwater, B. W. Dynamic-field devices for the ultrasonic manipulation of microparticles. Lab Chip 16, 2360–2375 (2016).
Wiklund, M., Green, R. & Ohlin, M. Acoustofluidics 14: Applications of acoustic streaming in microfluidic devices. Lab Chip 12, 2438–2451 (2012).
Yang, S. et al. Acoustic tweezers for high-throughput single-cell analysis. Nat. Protoc. 18, 2441–2458 (2023).
Yang, S. et al. Harmonic acoustics for dynamic and selective particle manipulation. Nat. Mater. 21, 540–546 (2022).
Wu, M. et al. Sound innovations for biofabrication and tissue engineering. Microsyst. Nanoeng. 10, 170 (2024).
Zhao, S. et al. Topological acoustofluidics. Nat. Mater. 24, 707–715 (2025).
Rufo, J., Cai, F., Friend, J., Wiklund, M. & Huang, T. J. Acoustofluidics for biomedical applications. Nat. Rev. Methods Primers 2, 30 (2022).
Rufo, J., Zhang, P., Zhong, R., Lee, L. P. & Huang, T. J. A sound approach to advancing healthcare systems: the future of biomedical acoustics. Nat. Commun. 13, 3459 (2022).
He, Y. et al. Acoustic technologies for the orchestration of cellular functions for therapeutic applications. Sci. Adv. 11, eadu4759 (2025).
Yang, S. et al. Acoustic tweezers for advancing precision biology and medicine. Nat. Rev. Methods Primers 5, 49 (2025).
Comfort, N. et al. Nanoparticle tracking analysis for the quantification and size determination of extracellular vesicles. J. Vis. Exp. 169, e62447 (2021).
Kim, J. et al. Comparison of EV characterization by commercial high-sensitivity flow cytometers and a custom single-molecule flow cytometer. J. Extracell. Vesicles 13, e12498 (2024).
Acknowledgements
We acknowledge support from the Shared Materials Instrumentation Facility (SMIF) at Duke University. We used ChatGPT (OpenAI) to help improve the clarity and readability of part of the manuscript after the initial draft was completed. We acknowledge support from the National Institutes of Health (R01HD103727, R01AG084098, R01GM141055, R01GM143439, R01GM145960, R01GM144417, U18TR003778 and UH3TR002978), and the National Science Foundation (CMMI-2104295). This publication includes data generated at the UC San Diego IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (no. S10 OD026929).
Author information
Authors and Affiliations
Contributions
J.X., L.P.L. and T.J.H. designed the research. J.X., B.L., A.G., C.C., J.P.N. and L.C.L. performed the research. J.X., B.L., A.G., C.C., S.Y., J.P.N. and L.C.L. analyzed data. J.X., B.L. and L.P.L. drew the figures. All authors wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.J.H. has cofounded a start-up company, Ascent Bio-Nano Technologies Inc., to commercialize technologies involving acoustofluidics and acoustic tweezers. J.P.N. is founder of Cellarcus Biosciences, which provides products and services for EV research. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Stephanie Descroix and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Xia, J. et al. ACS Nano 18, 22596–22607 (2024): https://doi.org/10.1021/acsnano.4c09692
Wu, M. et al. Proc. Natl Acad. Sci. USA 114, 10584–10589 (2017): https://doi.org/10.1073/pnas.1709210114
Wang, Z. et al. Microsyst. Nanoeng. 7, 20 (2021): https://doi.org/10.1038/s41378-021-00244-3
Shi, J. et al. Lab Chip 9, 3354–3359 (2009): https://doi.org/10.1039/B915113C
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2, Figs. 1–8 and Table 1.
Source data
Source Data Fig. 1 and Fig. 4
NTA results in Fig. 1d is provided. Vesicles flow cytometry results in Fig. 4f and 4g are provided. RNA sequencing results in Fig. 4h–j are provided.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xia, J., Lu, B., Yang, S. et al. Acoustic separation and isolation of viruses, small extracellular vesicles and other nanoscale bioparticles. Nat Protoc (2026). https://doi.org/10.1038/s41596-025-01286-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01286-x