Abstract
We present a dataset of plant hydraulic and structural traits imputed for 55,779 tree species based on TRY plant trait dataset observations and phylogenetic relationships. We collected plant trait values for maximum stomatal conductance (gsMAX), xylem pressure at 12%, 50%, and 88% conductance loss (P12, P50, P88), maximum observed rooting depth (rdMAX), photosynthetic Water Use Efficiency (WUE), maximum plant height (height), Specific Leaf Area (SLA), and leaf Nitrogen content (LeafN). We demonstrated that each of these traits exhibited remarkably large phylogenetic signals across all land plants. Based on the strength of this signal we then developed random forest (RF) models trained on TRY trait data to impute the traits of previously unstudied tree species using Phylogenetic Eigenvector Maps. We quantified imputed trait uncertainty by fitting RF model test dataset residuals to skew exponential power distributions accounting for heteroscedasticity, demonstrating encouraging lack of biases in the imputed dataset. The resulting dataset of imputed trait values can support global analyses of plant trait variations and species-level parameterization of earth systems models.
Similar content being viewed by others
Background & Summary
Hydraulic and structural traits define how plants uptake and transpire water from soils and groundwater, influencing ecosystem productivity, ecosystem resilience, and drought-induced mortality1,2,3. The traits of the plant species that cover landscapes determine the land surface energy balance, hydrologic partitioning (i.e., infiltration of precipitation versus surface runoff), and the degree to which subsurface water pools are connected to the atmosphere through transpiration4,5,6,7. Advances in process-based ecosystem modelling allow for the detailed representation of plant hydraulics in order to resolve the soil-plant-atmosphere-continuum which connects ecosystem water, nutrient, and energy fluxes with primary productivity8,9,10,11. These ecosystem models provide the opportunity to forecast earth system responses to both atmospheric and biological change12.
While the importance of these plant traits is well understood13 we lack trait measurements for most known tree species. A lack of direct trait observations to inform model parameterization has been part of the motivation for the compilation of global plant trait databases, such as the TRY Global Trait Database14,15. More than a decade into these efforts, a few traits are now reasonably well sampled globally, principally traits related to leaf economics such as leaf mass per area and leaf nitrogen content. However, even for these few well sampled traits, most traits have never been sampled for the vast majority of species globally (e.g. specific leaf area or SLA values exist for ~16,000 of Earth’s approximately ½ million land plants in TRY14). Observations of multiple traits in the same species are extremely rare, taken against the backdrop of global plant diversity, even for the simplest traits such as plant height and growth form14. For more difficult to measure physiological traits such as hydraulic traits, this data scarcity is even more dire. Models frequently forgo this complexity by representing vegetation with a small number of plant functional types, and therefore may be limited in their capacity to forecast earth systems processes16,17,18,19. As a result, there has been a call for creative efforts to parameterize the ‘functional types’ (discrete parameter sets that represent functional diversity in vegetation models), for example using evolutionary lineages to help guide the aggregation of trait values16.
Alternative methods exist for estimating plant traits beyond direct measurement in the field; however, each carries limitations. Remote sensing products can support estimating ecosystem-scale hydraulic traits20 with some advancement towards retrieving functional trait diversity from spectral signals21. Plant traits can also be inversely estimated through process-based ecosystem model fitting to species-level empirical field datasets (e.g., sapflux, xylem water isotopic compositions)22,23; however these measurements are resource intensive to collect and infrequently available. Given the limitations of current inverse approaches for estimating species-level hydraulic traits, a broad first order approximation of plant trait values could substantially advance ecosystem and earth systems modelling. Missing values in trait datasets can be imputed via methods such as Bayesian hierarchical probabilistic matrix factorization which can leverage the statistical structure of trait values, correlations among traits, and taxonomic relationships24,25; however these approaches have been tested primarily for highly sampled traits and rely on existing parallel measurements of other correlated traits. These approaches therefore may not satisfy the need for a tool that extrapolates to previously unstudied species.
Plant traits typically exhibit strong phylogenetic signals (i.e., more closely related species exhibit more similar trait syndromes than distantly related species)26,27,28,29,30, providing the opportunity to impute traits for previously unstudied species based on the relationship between functional traits and widely available phylogenetic data. We first performed a series of significance tests for phylogenetic signals in the hydraulic traits maximum stomatal conductance (gsMAX), xylem pressure at 12% (P12), 50% (P50), and 88% (P88) reduction in branch conductance, maximum rooting depth (rdMAX), water use efficiency (WUE), as well as the structural traits maximum plant height, specific leaf area (SLA), and leaf nitrogen composition per unit leaf mass (LeafN). We then imputed trait values for 55 K tree species based only on phylogenetic relationships and the TRY plant trait database15. This dataset of imputed values will support species-level ecosystem modelling and investigations of relationships between plant traits and environmental boundary conditions.
Methods
Plant trait phylogenetic signals
We collected plant trait values for maximum stomatal conductance (gsMAX), xylem pressure at 12%, 50%, and 88% conductance loss (P12, P50, and P88, respectively), maximum observed rooting depth (rdMAX), photosynthetic water use efficiency (assimilation/transpiration, or WUE), maximum plant height (height), Specific Leaf Area (SLA), and leaf nitrogen content per unit mass (LeafN) from the TRY database15. Plant trait records were filtered to remove values with TRY ErrorRisk values greater than 5 (indicating that the value is greater than five standard deviations from either the species-mean, genus-mean, family-mean or mean of all data for that trait, likely indicative of a data error) where ErrorRisk estimates were present, unflagged values that were likely data entry errors (e.g., negative stomatal conductance), and the over-representation of two crops (Coffea arabica and Glycine max). Documentation of TRY database filtering is provided in publicly available code attached to this work. Where multiple records existed for a single species, we computed the species median trait value. We validated each record name against World Flora Online (WFO), a comprehensive list of plant species31 with the R package ‘WorldFlora’. TRY species names that did not match WFO were corrected. Where corrections were not possible, observations were discarded. Validated plant species were mapped to a phylogeny using V.Phylomaker in the R package ‘V.Phylomaker2’32,33. Species not present in the backbone phylogeny were bound using ‘V.phylomaker2‘ under the scenario 3, which is the most commonly used approach. The scenario 3 methodology binds any new genus to an intermediate point of its family branch length and any species of an existing genus to the basal node of its genus. It varies from scenarios 1 and 2 as they bind any new tip to the genus or family basal node and to a random node within the genus or family, respectively33. The three scenarios have been compared in previous works, showing how scenarios 1 and 3 perform better and give similar results32. Therefore, we opted to use scenario 3. The resulting phylogenies contained the following unique species: gsMAX (n = 2,377), P12 (n = 387), P50 (n = 682), P88 (n = 436), rdMAX (n = 1,498), WUE (n = 317), height (n = 5,775), SLA (n = 12,595), and LeafN (n = 5,141).
Imputing plant traits using phylogenetic relationships requires first establishing that traits exhibit phylogenetic signals. We tested the hypothesis that each trait exhibited a significant phylogenetic signal with Pagel’s λ, which can be interpreted as a measure of the amount of variance explained by phylogenetic distances between species (ranging between 0 and 1)34, using 100 iterations as implemented in the R package ‘phytools’35. For this and all subsequent hypothesis tests we compared our p-values to ɑ thresholds of 0.1, 0.05, and 0.01. We also computed the fractions of trait variance explained by the phylogeny, VarPhylo, and their associated p-values30.
We acknowledge that species-level phylogenies may contain larger inaccuracies than deeper in the phylogenetic tree, especially when representing tropical taxa36. To assess the potential impact of such topological inaccuracies, we repeated this analysis for TRY traits with Pagel’s λ aggregated to the genus-level, pruning the species-level phylogeny keeping one species per genus (equivalent to a genus-level phylogeny). As will be demonstrated, phylogenetic signals maintained their significance, showing how most of the phylogenetic variance was explained by deep evolutionary divergences representing distances between well resolved high taxonomic ranks, in line with coarser taxonomic decomposition analyses of these same traits16. This verified that species-level phylogenetic patterns are not strongly affected by the phylogenetic distances within genera, which can contain a higher amount of error.
Estimation of species-level hydraulic and structural traits
To facilitate prediction of species-level hydraulic and structural traits, we repeated the above analysis; however, we retained all individual trait observation values (rather than collapsing all observations of each species to one median trait value). Phylogenies were constructed following the same approach. We then reduced these phylogenies to Phylogenetic Eigenvector Maps (PEM) which characterize the distances between species37. The original TRY trait observations were then joined to PEMs which could then serve as predictors of trait values.
We constructed all Random Forest (RF) models to predict trait values from PEMs with the R package ‘h2o’38. We then compared two methods for RF feature selection. First, using gsMAX, we trained the RF model on all PEMs. We then iteratively dropped the single PEM predictor with the lowest variable importance score and retrained the model. This process was repeated until RF performance significantly decreased when additional columns were removed. Second, we used a filter-based approach where we retained PEM predictors for model training that exhibited the strongest Spearman’s rank correlations with the observed trait values. RF model tests suggested that performance for the Spearman-based approach was similar for models retaining between 25 and 75 columns. We therefore used the 50 strongest rank-correlated columns. The two approaches to feature selection yielded similar RF performance. We selected the simpler filter-selection approach for imputing all plant traits.
RF models parameters included 300 trees, maximum depth of 50, and 8-fold cross validation. To estimate RF prediction uncertainty, the database was divided into training, validation, and test datasets based on 70%:15%:15% splits. The stopping condition used for training was Mean Squared Error. To estimate trait prediction performance, splits were developed by randomly sampling subsets of species such that all records each species occur only in one of the training, validation, or test datasets. We present four RF test dataset objective function values for each trait: Mean Absolute Scaled Error (MASE), Mean Absolute Error (MAE), R2, and Percent Bias (P-bias). All model metrics are computed only for the 15% of observations that were not used in model training/validation.
RF models using all TRY records for training and validation (i.e., no test hold out) were used to impute the trait values for tree species listed in the BCGI Global Tree Search dataset of 57,922 named species39. Validating and correcting tree species names in this list against WFO yielded 55,779 species names. TRY observations exist for the following fractions of species contained within the global tree list for the following traits: gsMAX (2.07%), P12 (0.52%), P50 (0.94%), P88 (0.60%), rdMAX (0.73%), WUE (0.33%), height (2.52%), SLA (10.19%), and LeafN (9.22%) of all species.
We compared the above approach to several parallel methodologies for imputing traits to provide context for the final dataset. We first compared using PEMs to Principal Coordinate Analysis (PCoA) as implemented in the R ‘ape’ package40. Next, we repeated the PEM-based analysis for P12, P50, and P88 records in the xylem functional trait database41 to test whether more curated (but smaller) hydraulic datasets yielded similar results. This dataset was filtered to include only stem samples from adult trees with S-shaped PLC curves.
Imputed trait residual characteristics and uncertainty bound estimation
The accuracy of imputed hydraulic and structural traits were quantified with RF test dataset residuals (i.e., e = predicted trait values - observed trait values). It was possible that RF trait residuals would be larger for tree species with greater documented within-species trait variations and for trees with fewer closely related species contained in the TRY database. We therefore hypothesized that RF residuals for all test datasets would exhibit significant phylogenetic signals. We tested for significant phylogenetic signals in model residuals with Pagel’s λ as described above. As will be demonstrated, model residuals were not significantly related to species-identity or phylogenetic relatedness for any traits. We therefore did not consider species identity in constructing statistical models of RF residuals.
Uncertainty bound estimates for each trait prediction were developed by fitting RF trait residual datasets to Skew Exponential Power (SEP) distributions with standard deviations accounting for residual heteroscedasticity42. Best-fit SEP parameters describing residual kurtosis, skew, and variance were estimated through Maximum Likelihood Estimation via 1e6 Monte Carlo simulations for each set of trait residuals. Fitted SEP distributions were then used to construct 50% confidence intervals for each imputed trait for ease of use, though we note that the provided SEP parameter values and code support construction of any confidence interval as well as Monte Carlo sampling of trait uncertainty.
Data Records
The global imputed trait dataset is publicly available on Zenodo43. The dataset consists of an R scripting language R Data Serialization (RDS) file, a Matlab MAT-file object, and an Excel spreadsheet (GlobalTrees_Traits_Median.xlsx), each containing median estimated trait values. The provided Skew Exponential Power (SEP) distribution parameters (Table 2) and median imputed trait values support the generation of random permutations of plant trait values for Monte Carlo simulations, (e.g. for parameter sensitivity analyses or forecast uncertainty using process-based vegetation models). Code to generate random permutations of plant traits from median values and SEP distribution parameters is available (see Code Availability).
Technical Validation
Plant trait phylogenetic signals
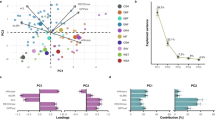
All median plant hydraulic, economic and structural trait values exhibited significant phylogenetic signals based on Pagel’s λ and VarPhylo at the ɑ < 0.01 threshold (Fig. 1). The phylogenetic dendrograms for maximum plant height, SLA, and LeafN are shown in Fig. 2. Genus-level analysis of phylogenetic signals yielded a similar result (Table 1). This result largely agrees with prior research demonstrating strong phylogenetic signals in plant hydraulic and structural traits26,27,29. The phylogenetic signal in all tested traits was highly statistically significant (based on both λ and VarPhylo). Phylogenetic variance was generally quite high (>65%) for all traits, with the exception of gsMAX and rdMAX
Estimation of species-level hydraulic and structural traits
Predicted plant hydraulic traits for the test datasets using PEMs demonstrated a reasonable predictive skill of the underlying RF models (Fig. 3). Mean Absolute Scaled Error (MASE) values for all test datasets were less than 1, indicating the RF models substantially outperformed the mean of the TRY database for each trait. Observed P-bias scores, with the exception of WUE, were all close to 0%, indicating that the RF models were mostly unbiased predictors of trait values. There also was no obvious dichotomy, either in observed phylogenetic signal nor RF model skill between the more classic leaf economics traits (SLA, Leaf N) and less well-sampled water use traits (P50, WUE), potentially supporting similar levels of phylogenetic conservatism among the traits that dictate carbon, water and nutrient strategies.
Random Forest (RF) test dataset model performance using PEMs for (a) maximum stomatal conductance (gsMAX), xylem pressure at (b) 12%, (c) 50%, and (d) 88% conductance loss (P12, P50, P88), (e) maximum observed rooting depth (RDMAX), (f) Water Use Efficiency (WUE), (g) height, (h) Specific Leaf Area (SLA), and (i) Leaf N content (Leaf N).
Trait values for P12 (Fig. 3b) were somewhat more poorly predicted than all other traits as measured by RF model R2 scores, despite this trait exhibiting a strong phylogenetic signal within TRY (Fig. 1). Imputed P12 values for some species are more negative than the predicted P50 value (Fig. 4a), an inconsistency that is largely absent between P50 and P88 (Fig. 4b). This further suggested high uncertainty in imputed P12 values relative to P50 and P88. Prior studies have noted that xylem pressures at turgor loss (often similar in magnitude and potentially mechanistically related to P12) can exhibit a weaker phylogenetic signal than P5027, which may explain the reduction in predictive skill. Alternatively, the substantial methodological uncertainty of hydraulic vulnerability curve measurements may make P12 or Pe (the point of initial air entry into xylem, often assumed to be near P12) inherently more difficult to measure than P50 across different methods. Alternatively, P12 may be negatively influenced by the composition of the TRY database. There is a disproportionate representation of conifers within TRY, though this is also true for P50 and P88 (Fig. 1). The distribution and few number of observed species for P12 in TRY may be limiting the computed PEMs from fully characterizing trait variations across the phylogeny.
We demonstrate that the PEM approach yields similar test dataset objective function values to a RF model trained on Principal Coordinate Analysis (PCoA) (Fig. 5) as implemented in the R ‘ape’ package40. RF model performance based on records in the xylem functional traits database, which is more curated and more easily screened but smaller than the TRY database, showed slightly improved prediction scores relative to TRY for P12, P50, and P88 (Fig. 6). Though this dataset shows promise for future use, we did not consider it further due to the small dataset size.
Random Forest (RF) test dataset model performance using PCoAs for (a) maximum stomatal conductance (gsMAX), xylem pressure at (b) 12%, (c) 50%, and (d) 88% conductance loss (P12, P50, P88), (e) maximum observed rooting depth (RDMAX), (f) Water Use Efficiency (WUE), (g) height, (h) specific leaf area (SLA), and (i) Leaf N content (Leaf N).
The intention of this dataset is to support global trait analyses and earth systems model forecasts that are by necessity climatic and ecological extrapolations. Our methodology intentionally excluded local environmental conditions from training despite the promise that these approaches have shown as hindcasting tools. By excluding this information, we produced a dataset of imputed traits and their associated uncertainties that reflects the broadest range of environmental conditions possible. The trait dataset conditioned only on phylogenies is therefore more robust with respect to the broad need for ecosystem model parameterizations that are climate-transferable19,44,45.
Imputed trait residual characteristics and uncertainty bound estimation
RF model residuals for all traits did not exhibit significant phylogenetic signals at the α < 0.1 threshold (Fig. 7). Residuals for WUE showed a high λ value, but the result was not significant possibly due to the relatively smaller dataset size. We expected that issues of data sparsity, non-random sampling of the phylogeny for some traits, and other issues with the training data would result in phylogenetically structured model errors. However, the RF models apparently captured the phylogenetic structure of the data extremely well for all traits. This result suggested that RF performance did not vary significantly with tree species identity. We therefore did not consider species identity in constructing statistical models of plant trait residuals.
Phylogenies of Random Forest (RF) model residuals for (a) maximum stomatal conductance (gsMAX), xylem pressure at (b) 12%, (c) 50%, and (d) 88% conductance loss (P12, P50, P88), (e) maximum observed rooting depth (RDMAX), (f) Water Use Efficiency (WUE), (g) height, (h) Specific Leaf Area (SLA), and (i) Leaf N content (Leaf N), showing Pagel’s λ and p-values in parentheses.
RF model residuals were well described by Skew Exponential Power (SEP) distributions accounting for heteroscedasticity (Figs. 8, 9; Table 2). All trait residuals exhibited very limited skew (similar to P-bias scores near 0%), further demonstrating that the RF models were unbiased predictors. All traits exhibited some degree of heteroscedasticity where residual variance increased with the magnitude of the trait being predicted (Fig. 8, Table 2).
(1) Scatter plots of Random Forest imputed traits versus test dataset residuals showing heteroscedasticity and (2) histograms of test dataset residuals for (a) maximum stomatal conductance (gsMAX), xylem pressure at (b) 12%, (c) 50%, and (d) 88% conductance loss (P12, P50, P88), (e) maximum observed rooting depth (RDMAX), (f) Water Use Efficiency (WUE), (g) height, (h) Specific Leaf Area (SLA), and (i) Leaf N content (Leaf N).
The cause of the observed residual heteroscedasticity could potentially be explained by trait measurement errors within TRY, where the magnitude of measurement biases scale with the measurement being taken. For example, tree height uncertainty measurements are often expressed as a percentage46, implying that height uncertainty increases linearly as a function of height. Another possibility is that plants may tend to evolve similar strategies for survival27, resulting in few plant records within TRY that represent extreme trait values. The underrepresentation of extremal trait values in the training datasets may have limited the ability of the RF models to learn where large magnitude trait values are likely to occur across the phylogeny, resulting in residuals that scale in magnitude with trait values. Given that underlying traits exhibited strong phylogenetic signals (Fig. 1, Table 1) but that test dataset residuals did not exhibit significant phylogenetic signals (Fig. 7) this explanation may be less likely.
Code availability
Code used in the preparation of this work is available at the following link: https://github.com/jknigh0813/PlantTraitDatabase.
Change history
05 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41597-025-05244-w
References
Anderegg, W. R. L. et al. Hydraulic diversity of forests regulates ecosystem resilience during drought. Nature 561, 538–541 (2018).
Brodribb, T. J., Powers, J., Cochard, H. & Choat, B. Hanging by a thread? Forests and drought. Science 368, 261–266 (2020).
Sanchez-Martinez, P. et al. Increased hydraulic risk in assemblages of woody plant species predicts spatial patterns of drought-induced mortality. Nat Ecol Evol 7, 1620–1632 (2023).
Evaristo, J. & McDonnell, J. J. Prevalence and magnitude of groundwater use by vegetation: a global stable isotope meta-analysis. Sci Rep 7, 44110 (2017).
Fan, Y., Miguez-Macho, G., Jobbágy, E. G., Jackson, R. B. & Otero-Casal, C. Hydrologic regulation of plant rooting depth. Proceedings of the National Academy of Sciences 114, 10572–10577 (2017).
Good, S. P., Noone, D. & Bowen, G. Hydrologic connectivity constrains partitioning of global terrestrial water fluxes. Science 349, 175–177 (2015).
Valverde-Barrantes, O. J., Freschet, G. T., Roumet, C. & Blackwood, C. B. A worldview of root traits: the influence of ancestry, growth form, climate and mycorrhizal association on the functional trait variation of fine-root tissues in seed plants. New Phytologist 215, 1562–1573 (2017).
Li, L. et al. Representation of Plant Hydraulics in the Noah-MP Land Surface Model: Model Development and Multiscale Evaluation. Journal of Advances in Modeling Earth Systems 13, e2020MS002214 (2021).
Ruffault, J., Pimont, F., Cochard, H., Dupuy, J.-L. & Martin-StPaul, N. SurEau-Ecos v2.0: a trait-based plant hydraulics model for simulations of plant water status and drought-induced mortality at the ecosystem level. Geoscientific Model Development 15, 5593–5626 (2022).
Kennedy, D. et al. Implementing Plant Hydraulics in the Community Land Model, Version 5. Journal of Advances in Modeling Earth Systems 11, 485–513 (2019).
Knighton, J. et al. Using isotopes to incorporate tree water storage and mixing dynamics into a distributed ecohydrologic modelling framework. Ecohydrology 13, e2201 (2020).
Stephens, C. M., Lall, U., Johnson, F. M. & Marshall, L. A. Landscape changes and their hydrologic effects: Interactions and feedbacks across scales. Earth-Science Reviews 212, 103466 (2021).
Anderegg, W. R. L. & Venturas, M. D. Plant hydraulics play a critical role in Earth system fluxes. New Phytologist 226, 1535–1538 (2020).
Kattge, J. et al. TRY – a global database of plant traits. Global Change Biology 17, 2905–2935 (2011).
Kattge, J. et al. TRY plant trait database – enhanced coverage and open access. Global Change Biology 26, 119–188 (2020).
Anderegg, L. D. L. et al. Representing plant diversity in land models: An evolutionary approach to make “Functional Types” more functional. Global Change Biology 28, 2541–2554 (2022).
Matheny, A. M., Mirfenderesgi, G. & Bohrer, G. Trait-based representation of hydrological functional properties of plants in weather and ecosystem models. Plant Diversity 39, 1–12 (2017).
Peaucelle, M. et al. Covariations between plant functional traits emerge from constraining parameterization of a terrestrial biosphere model. Global Ecology and Biogeography 28, 1351–1365 (2019).
Adams, H. D. et al. Climate-Induced Tree Mortality: Earth System Consequences. Eos, Transactions American Geophysical Union 91, 153–154 (2010).
Liu, Y., Holtzman, N. M. & Konings, A. G. Global ecosystem-scale plant hydraulic traits retrieved using model–data fusion. Hydrology and Earth System Sciences 25, 2399–2417 (2021).
Schweiger, A. K. et al. How to predict plant functional types using imaging spectroscopy: linking vegetation community traits, plant functional types and spectral response. Methods in Ecology and Evolution 8, 86–95 (2017).
Li, K., Kuppel, S. & Knighton, J. Parameterizing Vegetation Traits With a Process-Based Ecohydrological Model and Xylem Water Isotopic Observations. Journal of Advances in Modeling Earth Systems 15, e2022MS003263 (2023).
De Deurwaerder, H. P. T. et al. Causes and consequences of pronounced variation in the isotope composition of plant xylem water. Biogeosciences 17, 4853–4870 (2020).
Shan, H. et al. Gap filling in the plant kingdom: trait prediction using hierarchical probabilistic matrix factorization. in Proceedings of the 29th International Coference on International Conference on Machine Learning 331–338 (Omnipress, Madison, WI, USA, 2012).
Joswig, J. S. et al. Imputing missing data in plant traits: A guide to improve gap-filling. Global Ecology and Biogeography 32, 1395–1408 (2023).
Knighton, J., Fricke, E., Evaristo, J., de Boer, H. J. & Wassen, M. J. Phylogenetic Underpinning of Groundwater Use by Trees. Geophysical Research Letters 48, e2021GL093858 (2021).
Sanchez-Martinez, P., Martínez-Vilalta, J., Dexter, K. G., Segovia, R. A. & Mencuccini, M. Adaptation and coordinated evolution of plant hydraulic traits. Ecology Letters 23, 1599–1610 (2020).
Swenson, N. G. Phylogenetic imputation of plant functional trait databases. Ecography 37, 105–110 (2014).
Ávila-Lovera, E., Winter, K. & Goldsmith, G. R. Evidence for phylogenetic signal and correlated evolution in plant–water relation traits. New Phytologist 237, 392–407 (2023).
Sanchez-Martinez, P. et al. A framework to study and predict functional trait syndromes using phylogenetic and environmental data. Methods in Ecology and Evolution 15, 666–681 (2024).
Borsch, T. et al. World Flora Online: Placing taxonomists at the heart of a definitive and comprehensive global resource on the world’s plants. TAXON 69, 1311–1341 (2020).
Jin, Y. & Qian, H. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography 42, 1353–1359 (2019).
Jin, Y. & Qian, H. V.PhyloMaker2: An updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Diversity 44, 335–339 (2022).
Pagel, M. Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3, 217–223 (2012).
Baker, T. R. et al. Maximising Synergy among Tropical Plant Systematists, Ecologists, and Evolutionary Biologists. Trends in Ecology & Evolution 32, 258–267 (2017).
Guénard, G., Legendre, P. & Peres-Neto, P. Phylogenetic eigenvector maps: a framework to model and predict species traits. Methods in Ecology and Evolution 4, 1120–1131 (2013).
LeDell, E. R Interface for the H2O Scalable Machine Learning Platform https://docs.h2o.ai/h2o/latest-stable/h2o-r/docs/index.html (2020).
Beech, E., Rivers, M., Oldfield, S. & Smith, P. P. GlobalTreeSearch: The first complete global database of tree species and country distributions. Journal of Sustainable Forestry 36, 454–489 (2017).
GOWER, J. C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338 (1966).
Choat, B. et al. Global convergence in the vulnerability of forests to drought. Nature 491, 752–755 (2012).
Schoups, G. & Vrugt, J. A. A formal likelihood function for parameter and predictive inference of hydrologic models with correlated, heteroscedastic, and non-Gaussian errors. Water Resources Research 46 (2010).
Knighton, J., Sanchez-Martinez, P. & Anderegg, L. A Globally Comprehensive Database of Tree Hydraulic and Structural Traits Imputed from Phylogenetic Relationships. Zenodo https://doi.org/10.5281/zenodo.15009207 (2025).
Broderick, C., Matthews, T., Wilby, R. L., Bastola, S. & Murphy, C. Transferability of hydrological models and ensemble averaging methods between contrasting climatic periods. Water Resources Research 52, 8343–8373 (2016).
Rogger, M. et al. Land use change impacts on floods at the catchment scale: Challenges and opportunities for future research. Water Resources Research 53, 5209–5219 (2017).
Larjavaara, M. & Muller-Landau, H. C. Measuring tree height: a quantitative comparison of two common field methods in a moist tropical forest. Methods in Ecology and Evolution 4, 793–801 (2013).
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant Nos. 2243263, 2003205, 20230833 and 2216855 and Renewable Energy, Natural Resources, and Environment: Agroecosystem Management Grant no. GRANT13398847 / project accession no. 1027642 from the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Contributions
J.K. - conception, data aggregation, analysis, quality control, manuscript preparation. P.S.M. - analysis, quality control, manuscript preparation. L.A. - conception, quality control, manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Knighton, J., Sanchez-Martinez, P. & Anderegg, L. A global dataset of tree hydraulic and structural traits imputed from phylogenetic relationships. Sci Data 11, 1336 (2024). https://doi.org/10.1038/s41597-024-04254-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-024-04254-4