Abstract
Procypris rabaudi, commonly known as the rock carp, is an endemic economic fish in the middle-upper reaches of the Yangtze River. To enhance the understanding the biology of rack carps, a high-quality reference genome is required in different areas of study. Here, we generated the first telomere-to-telomere genome assembly and annotation of the rock carp, which spans 1.64 Gb with a contig N50 of 32.36 Mb. Hi-C assembly suggested that 99.83% sequences were positioned to 50 pseudo-chromosomes. Notably, 43 chromosomes were assembled without any gap. Through the integration of homologous-based predictions and RNA-sequencing technology, we identified 44,402 protein-coding genes, with 43,663 of them (98.3%) predicted to be functional. Furthermore, our assembled genome achieved 98.1% BUSCO completeness. This work improves the quality of the rock carp genome and provides valuable foundation for the future studies of genomics, biology and the fishery resources breeding.
Similar content being viewed by others
Background & Summary
The rock carp, Procypris rabaudi (Tchang, 1930) (Fig. 1A), belongs to the Cyprinidae family and is an important endemic economic species of the middle-upper reaches of the Yangtze River, including the Jinsha River, Jialing River, Minjiang River, Chishui River and Tuojiang River1. Rock carp is a benthic fish that lives mainly in deep waters with rocky substrate, and produces adhesive eggs to attach to the gravel substrate at the bottom during the spawning season2. Since 1970s, the populations of rock carp have declined sharply due to anthropogenic influence like pollution, overfishing, among others3. At present, the rock carp is listed as the second-class aquatic animals in the National List of Key Protected Wild Animals of China. In recent decades, the artificial propagation and release has been applied to restore the populations of rock carp4. The great efforts have been adopting to protect and recover their wild stocks in the fields. Moreover, molecular genetics provides critical reference for phylogeographic research and breeding, but this relies on sequencing effort and genome assembly.
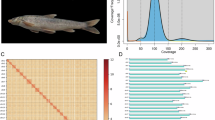
(A) Genomic landscape of the rock carp. The rings, from the outermost to the innermost layer, represent the chromosomes of the P. rabaudi genome (a), gene density (b), GC density (c), short reads density (d), long reads density (e), outer ring shows the homology SNPs density, inner ring shows the heterozygosis SNPs density (f), outer ring shows the homology InDel density, inner ring shows the heterozygosis InDel density (g). The completed mapped BUSCO genes density, single-copy BUSCOs is blue and duplicated BUSCOs is red (a-g). The analysis was conducted using 50-kb genomic windows. (B) Chromosomal Hi-C heatmap of the P. rabaudi genome assembly.
With the development of sequencing technologies, the sequencing cost has dramatically reduced5. This further promotes the application of high-quality genomes in basic biology. A study on the origin and subsequent subgenome evolution patterns has published twenty-one cyprinids genomes including the rock carp6,7. Although the genome assembly of rock carp have been available, there is still space for improvement in genome assembly and annotation. In-depth research of the rock carp is still be constrained. Presently, the assembly of telomere-to-telomere (T2T) gap-free genome have been possible for the advances in sequencing and assembly technology. In bony fish, T2T genome assemblies of several important species have been reported and provided new insight, like the zig-zag eel T2T genome revealed the origin and evolutionary of its sex chromosome8, the Chinese sea bass T2T genome provided reference for further analysis of its genome structure and mining of breeding genes for disease resistance9. For the rock carp, its genome is relatively complex as a tetraploid species, so a higher quality genome is necessary to analyze its genome structure. In addition, this will further promote the genetic breeding, disease resistance research and resource conservation of the rock carp. Herin, we aim to assemble the genome of P. rabaudi to the T2T level, and this will provide a high-quality reference for the in-depth study of this species.
In this study, we generated Pacific Biosciences (PacBio), High-fdelity (HiFi) long-reads, ultra-long Oxford Nanopore (ONT) and Hi-C sequencing reads for P. rabaudi T2T genome assembly. Our genome assembly improves the previous chromosome-level rock carp genome and provides significant genomic resources of evolutionary and breeding research.
Methods
Sample collection and sequencing procedures
Adult female specimens of Procypris rabaudi were sourced from a fish farm located in Chongqing, China (coordinates: 122.212 E, 29.979 N). The research protocol was approved by the Animal Care and Use Committee at the Fishery College of Southwest University. Muscle and blood tissue was specifically chosen for DNA and ultra-long ONT extraction and sequencing respectively. Various tissues, including muscle, brain, blood, skin, liver, gonad and skull were harvested and stored for subsequent RNA isolation for genome annotation. A PacBio HiFi-read library, featuring insert sizes ranging from 10 to 40 kb, was prepared using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, USA) and sequenced on the PacBio Sequel II platform. For ONT ultra-long reads, an SDS-based extraction method was employed. Additionally, a Hi-C short-read library was constructed from purified DNA following the protocol established by Belton et al., and sequencing was conducted on the Illumina Novaseq 6000 platform10. The sequencing yielded 53.20 Gb of PacBio HiFi long-reads with an N50 of 19.11 kb, 20.21 Gb of ONT ultra-long reads with an N50 of 100 kb, and 142.59 Gb of clean Hi-C reads for genome assembly (refer to Table 1).
For genome annotation, RNA was extracted from all collected tissues. Total RNA was isolated using TRIzol reagent (Invitrogen, MA, USA) and processed into an RNA-seq library with the NEBNext® Ultra™ RNA Library Prep Kit (NEB, USA). Sequencing was performed on the Illumina Novaseq 6000 platform.
Genome assembly and gap filling
The initial genome assembly was conducted using HiFiasm (v0.16.0), integrating HiFi data, ONT ultra-long reads, and Hi-C sequencing data11. This resulted in a draft genome of 1.65 Gb with a contig N50 of 29.64 Mb (see Table 2). Chromosome-level assembly was achieved using Hi-C data. Clean Hi-C reads were aligned to the contig-level genome using Bowtie2 (v2.3.4.3), producing 227.07 million uniquely mapped paired-end reads, with 49.22% being valid pairs (Tables S1, S2)12. The 3D-DNA pipeline (v180922) and JuiceBox (v1.11.08) were utilized to calculate chromosomal interaction frequencies and correct scaffolding errors, respectively13,14. This process yielded 50 pseudo-chromosomes covering 99.83% of the genome, with a contig N50 of 28.65 Mb (Fig. 1B).
To achieve a near-T2T genome, ONT ultra-long reads were mapped to the pseudo-chromosomes using minimap215. TGS-GapCloser was used for gap filling and 3 rounds genome correction were performed by Pilon, resulting in a 1.64 Gb genome with a contig N50 of 32.36 Mb and 43 fully assembled chromosomes (Fig. 2, Table S3)16,17.
Telomere and centromeric regions analysis
The quarTeT (v1.1.4) software was used in telomere and centromere analysis18. Telomeres were identified by scanning the genome for the TTAGGG/CCCTAA motif, with at least four repetitions required for recognition. Based on the annotation results of genome and the identification results of quarTeT software, regions with dense distribution of Satellite and TandemRepeat were considered as candidate centromere regions. The estimated telomeres and centromere regions were obtained as shown in Fig. 2, Table S3.
Repetitive sequence analysis
Repetitive sequences, including tandem and interspersed repeats, were identified using de novo prediction and homology-based methods. A repeat library was constructed using RepeatModeler (open-1.0.11) and LTR-FINDER_parallel (v1.0.7)19,20. Tandem repeats were detected with TRF (v4.09), while RepeatMasker (open-4.0.9) and RepeatProteinMask were used for homology-based predictions21,22. In total, 0.79 Gb of repetitive sequences, accounting for 48.34% of the genome, were identified, including 28.73% DNA elements, 6.09% LINEs, and 6.04% LTRs (Table 3). And the information of repetitive sequences can be founded in Table S4.
Protein-coding gene annotation
The repeat-masked genome was subjected to ab initio gene prediction using AUGUSTUS (v3.3.2), Genscan (v1.0), and GlimmerHMM (v3.0.4)23,24,25. GeneWise (v2.4.1) was used for precise protein mapping and splice site identification26. RNA-seq reads were aligned to the genome using HISAT2 (v2.2.1), and transcripts were assembled with StringTie (v2.2.0) and PASA (v2.3.2)27,28,29. MAKER2 (v2.31.10) and HiFAP integrated these predictions, resulting in 44,402 protein-coding genes (Table 4)30. Comparative genomics with related species (Carassius carassius, Carassius gibelio, Cirrhinus molitorella, and Cyprinus carpio) was performed using TBLASTN (e-value ≤ 1e-5) to identify protein-coding regions31. Gene structures were compared with homologous species (Fig. 3).
Functional annotation of protein-coding genes was performed using InterPro, GO, KEGG, SwissProt, TrEMBL, TF, Pfam, NR, and KOG databases32,33,34,35,36. InterProScan (v5.61–93.0) was employed to annotate conserved domains and motifs, with 98.34% (43,663) of genes functionally annotated (Table 5)37.
Non-coding gene identification
The tRNA sequences were predicted using tRNAscan-SE (v1.3.1), while rRNA genes were identified via BLASTN. miRNA and snRNA sequences were predicted using INFERNAL based on the Rfam database (v14.8)38,39. Results are summarized in Table 6.
Data Records
All sequencing data from three sequencing platforms and the assembled genome have been uploaded to the NCBI SRA database and could be accessed with BioProject number PRJNA117582740,41,42,43,44,45. The genome annotation files could be founded in figshare: https://doi.org/10.6084/m9.figshare.2858838246.
Technical Validation
DNA and RNA quality assessment
Prior to sequencing, DNA and RNA quality (OD260/280 and OD260/230 ratios) and concentration were measured using the NanoDrop 2000 Spectrophotometer and Qubit 3.0 Fluorometer. Sample integrity was confirmed via agarose gel electrophoresis and the Agilent 2100 Bioanalyzer.
Genome assembly quality evaluation
Short reads were aligned to the assembled genome using BWA (v0.7.17 -r 1188), while HiFi and ONT ultra-long reads were mapped with Minimap2 (v2.24_x64-linux)15,47. Mapping rates were 99.88% for short reads and 99.99% for HiFi and ONT reads (Tables S5, S6). BUSCO (v5.4.3) analysis based on the actinopterygii_odb10 database indicated that 98.1% of 3571 single-copy orthologs were complete (Table 7)48.
Code availability
No specific code was used in this study. The standard bioinformatic tools were used for data analysis. Furthermore, the parameter setting of the bioinformatics tools was performed in accordance with the manual and protocols and described in the Methods Section.
References
Wang, T. et al. Life history traits and implications for conservation of rock carp Procypris rabaudi Tchang, an endemic fish in the upper Yangtze River. Fish. Sci. 81, 515–523 (2015).
Probst, W. et al. Rearing of Procypris rabaudi during early life‐history stages. J. Appl. Ichthyol. 22, 530–535 (2006).
Yang, L., Mayden, R. L. & Cai, Y. Z. Threatened fishes of the world: Procypris rabaudi (Tchang, 1930) (Cyprinidae). Environ. Biol. Fishes. 84, 275–276 (2009).
Zhang, X. et al. Study on the genetic structure of wild and hatchery populations of Procypris rabaudi Tchang, an endemic fish in the upper Yangtze River. Fish. Res. 245, 106134 (2002).
Jarvis, E. D. et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 346, 1320–1331 (2014).
Xu, M. R. X. et al. Maternal dominance contributes to subgenome differentiation in allopolyploid fishes. Nat. Commun. 14, 8357 (2023).
Database Resources of the National Genomics Data Center. China National Center for Bioinformation in 2022. Nucleic Acids Res. 50, D27–D38 (2022).
Xue, L. Z. et al. Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22, 203 (2021).
Sun, Z. L. et al. Telomere-to-telomere gapless genome assembly of the Chinese sea bass (Lateolabrax maculatus). Sci. Data 11, 175 (2024).
Belton, J. M. et al. Hi–C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Xu, M. Y. et al. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. Gigascience 9, giaa094 (2020).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one 9, e112963 (2014).
Lin, Y. Z. et al. quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic Res. 8, uhad127 (2023).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Ou, S. & Jiang, N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mobile DNA 10, 1–3 (2019).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Birney, E., Clamp, M. & Durbin, R. GeneWise and GenomeWise. Genome Res. 14, 988–995 (2004).
Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 905–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295 (2015).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 1–14 (2011).
McGinnis, S. & Madden, T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 (2004).
Apweiler, R. et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Finn, R. D. et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45, D190–D199 (2017).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014).
Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 1–14 (2003).
Bairoch, A. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 (2003).
Zdobnov, E. M. & Apweiler, R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 (2001).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124 (2005).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31066338 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31066339 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31066340 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31066341 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR31066342 (2024).
He, W. Procypris rabaudi isolate WH-2025, whole genome shotgun sequencing project. Genbank. https://identifiers.org/ncbi/insdc:JBMMEF000000000.1 (2025).
He, W. Genome annotation of Procypris rabaudi. figshare https://doi.org/10.6084/m9.figshare.28588382.v1.
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Acknowledgements
This research was funded by National Key Research and Development Program of China (2022YFD1201601), National Natural Science Foundation of China (32071651), Fundamental Research Funds for the Central Universities (SWU-XDJH202317), Technical Innovation and Application Development Project of Chongqing (CSTB2023TIADLDX0001), The Earmarked fund for CARS (CARS-49).
Author information
Authors and Affiliations
Contributions
W.P.H. designed this project. X.L.L. collected the samples. Q.L. analyzed the data. X.L.H. wrote the manuscript and L.H.X. revised the manuscript. S.Q.S., W.Z.Y. and W.P.H revised the manuscript and gave important advises to this work. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41597_2025_5066_MOESM1_ESM.docx
Supplementary information of the telomere-to-telomere genome assembly and annotation of the rock carp (Procypris rabaudi)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Han, X., Li, X., Xu, L. et al. The telomere-to-telomere genome assembly and annotation of the rock carp (Procypris rabaudi). Sci Data 12, 781 (2025). https://doi.org/10.1038/s41597-025-05066-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05066-w
This article is cited by
-
The haplotype-resolved and near telomere-to-telomere genome assembly for Hypsebarbus vernayi
Scientific Data (2025)