Abstract
Surface electromyography (sEMG) records muscle electrical signals and reflects neuromuscular physiological behaviors. Recently, high-density sEMG (HD-sEMG), which allows non-invasive identification of motor unit action potential trains (MUAPTs) and direct access to underlining neural drive derived from the spinal cord, becomes a research hotspot. However, datasets comprising HD-sEMG signals remain limited, especially for multi-day conditions, leading to the lack of long-term investigation of motor neuron activities. This paper presents a 320-channel HD-sEMG dataset, CEMHSEY (ConsecutivE Multi-day High-density Surface ElectromyographY), recorded from forearm muscles and across 11 consecutive days. The dataset consists of two sub-datasets as: an isometric contraction dataset containing 13 subjects performing 7 grasps under 3 different contraction force levels (named GRASP) and a hand gesture dataset with 6 subjects performing 11 hand gestures (named GESTURE). The dataset was validated with the usability of force regression, hand gesture recognition, and motor unit decoding. In addition, the multi-day data provide support for developing robust human-machine interfaces as well as analyzing neuromuscular modulation.
Similar content being viewed by others
Background & Summary
Surface electromyography (sEMG), recording electrical signals of muscles, contains information on motor unit discharge patterns and plays a pivotal role in constructing a non-invasive neural interface for bio-robotics1,2. Such an interface can function as a generalized human-machine interface (HMI) with diverse potential applications3. In the medical and healthcare domains, it facilitates neurorehabilitation and supervision of muscle physiological state, while in non-medical contexts, it enables interaction with multifunctional limb prostheses and other activated bio-robotics4,5,6. Recently, high-density sEMG (HD-sEMG) with multiple channels covering a certain skin area provides a wealth of physiological and anatomical information about the underlying muscles and allows decomposition into the motor unit action potential trains (MUAPTs), which identifies the individual motor unit features and their changes in pathologies.
Although HD-sEMG showcases promising prospects, there still exists a persistent gap between academic research and the actual industrial or medical applications, especially the lack of robust algorithms against daily non-stationary factors. Traditional control algorithms based on sEMG assume that features extracted from sEMG signals are sufficiently stationary, which is acceptable in the lab where short-term trials are conducted7. However, in real-life scenarios, the limb is susceptible to cross-day physical and physiological changes due to sweating, swelling, and fatigue. It is one of the reasons that a number of laboratory HD-sEMG-controlled prostheses models with high performance and recognition accuracy have been reported in previous studies, while commercial products exhibit poorer performance in daily activities of living compared to lab circumstances8. Cross-day non-stationary factors skin impedance, muscle physiological state, and electrode shift significantly affect the classification accuracy of the control algorithm2. An effective approach to enhance the performance of classification models and increase their robustness to scenario changes is referred to as data augmentation, which means readapting the trained model to additional data obtained under various conditions, e.g., multiple days9. This approach is effective in mitigating cross-day interference, yet additional multi-day HD-sEMG data is needed.
Further insight into adaptations of neuromuscular physiological state also requires HD-sEMG data acquired across multiple days. Much more than serving as robust input for HMI, MUAPTs decomposed from HD-sEMG provide rich information on motor unit discharging patterns and reflect spinal motor neuron activities. Some freely accessible sEMG datasets (shown in Table 1) provide technically validated experimental data for researchers. SEEDS10 contains within-day HD-sEMG data. GRABMyo11 contains sEMG data from 3 discontinuous days. CapgMyo12 contains HD-sEMG data from 2 nonadjacent days with an interval of more than one week, while Hyser13 contains HD-sEMG data from 2 adjacent days. However, none of them include consecutive multi-day HD-sEMG data, which serves as a tool to monitor neuron physiological state with a more precise temporal scale.
To fill in the blank of continuous multi-day HD-sEMG data in open-access datasets, we present a publicly available dataset with multiple channel data acquisition setup under consecutive multi-day experiment protocol. The dataset, named CEMHSEY, consists of two sub-datasets, namely GRASP and GESTURE. GRASP sub-dataset contains 11-day repetitions of 320-channel HD-sEMG data of the right forearm collected from 13 non-disabled participants performing 7 hand grasps, including cylindrical grasp (Cyl), lumbrical grasp (Lum), and oblique palmar grasp (Obl), as well as simultaneous hand grip force data corresponding to each grasp task. GESTURE sub-dataset contains 11-days repetitions of 320-channel HD-sEMG data of the right forearm collected from 6 non-disabled participants performing 11 hand gestures, including hand close (HC), hand open (HO), wrist flexion (WF). There are totally 6006 trials (13 participants × 11 days × 7 sessions × 3 tasks × 2 trials) in GRASP and 330 trials (6 participants × 11 days × 5trials) in GESTURE.
We believe that the dataset CEMHSEY presented in this paper will be useful to researchers to facilitate the research process concerning cross-day EMG-Force regression, hand gesture recognition, and neural drive extraction. Hopefully, it can contribute to the study of cross-day neurophysiological investigation, robust HMI, and algorithm development and validation.
Method
Participant
The data was collected from 14 non-disabled volunteers without any skeleton defect or neuromuscular diseases (13 males, 1 female). 13 subjects aged 19–31 years (21.46 ± 3.57 years) participated in GRASP experiment and 6 subjects aged 19–26 years (20.83 ± 2.56 years) participated in GESTURE experiment. 5 of them attended both experiments.
Volunteers were recruited via an online non-profit platform of Shanghai Jiao Tong University. Candidates who met the experimental conditions and schedule were selected for experiments. All participants had signed the informed consents before the experiment. The experiment conforms to the Declaration of Helsinki and was approved by the academic ethics committee of Shanghai Jiao Tong University (approval number: E20240248I).
Setup for data acquisition
High-density surface electromyography data acquisition
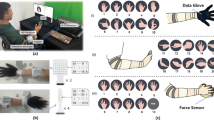
HD-sEMG signal was amplified with a gain of 150 and sampled at 2048 Hz with a 16-bit resolution and filtered in the monopolar mode through a first-order high pass filter with a cut-off frequency at 10 Hz as well as a first-order low pass filter with a cut-off frequency at 4400 Hz (Quattrocento, OT Bioelettronica, Italy). The arrangement of electrodes contains three 8 × 8 HD-sEMG electrode arrays (64 channels, GR10MM0808, OT Bioelettronica, Italy) with an inter-electrode distance of 10 mm in both horizontal and vertical directions and two 5 × 13 HD-sEMG electrode arrays (64 channels, GR08MM1305, OT Bioelettronica, Italy) with an inter-electrode distance of 8 mm in both horizontal and vertical directions. In total, there are 320 channels. The diameter of the electrode is 3 mm. Three 8 × 8 electrode grids were mounted around the proximal third of the right forearm and two 5 × 13 electrode grids were around the distal third of the right forearm (Fig. 1(b)). Sometimes, grid overlap due to the subject’s small forearm circumference was inevitable in the data acquisition process.
(a) Setup for data acquisition: the subject was seated comfortably in front of a graphical user interface (GUI) on the screen, performing different grasps or gestures as instructed by the GUI. In GRASP experiment (i), the right arm of the subject was supported by an arm holder. In GESTURE experiment (ii), the right arm of the subject was hanging naturally in no contact with the body. (b) Electrode grids placement on the subject’s right arm for recording HD-sEMG, same for both experiments. Each grid has 64 channels. The red figures indicate the grid number. For example, grid 1 corresponds to channel 1–64, and grid 5 corresponds to channel 257–320. (c), (d) The alignment of channels on grid 1, 2, and 3 (d) with 10 mm inter-electrode distance, and on grid 4 and 5 (e) with 8 mm inter-electrode distance.
Once the overlap occurred, the experimenter would maintain the overlap sequence consistent through the whole experiment period. For example, if part of Electrode array 3 was stacked on Electrode array 1 on the first day, then this situation should be maintained during the following ten days. On the first day of the whole experiment period, the volunteer used an exfoliating scrub to clean the skin. Before electrode attachment, an alcohol pad was used to clean the experimental region. After the daily experiment was over, the experimenter would outline the profile of each electrode array before taking down the electrodes with a marker to indicate their placement position, which helped them reposition electrodes for the next day. For GRASP and GESTURE sub-experiments, the electrodes would stay on the skin for approximately 40 minutes each day.
Force data acquisition
Force data was sampled at 200 Hz by a hand grip dynamometer (G200, Biometrics Ltd, United Kingdom) or a pinch dynamometer (P200, Biometrics Ltd, United Kingdom). Which force measuring apparatus was employed depended on which grasp the participant was performing. In the GRASP experiment, the hand grip dynamometer was used for the first four grasps, while the pinch dynamometer was used for the remaining three grasps.
GRASP sub-experiment would acquire HD-sEMG data as well as the force data simultaneously, and GESTURE sub-experiment would only acquire HD-sEMG data.
Experiment protocol
GRASP sub-experiment
The whole experiment for each participant lasted for 11 days. The volunteer was seated in front of the laptop screen, with the right elbow laid on an arm support and the forearm relaxed (Fig. 1(a)(i)). Each day, the subject would perform 7 sessions (grasps) of activity of daily living using their right hand: 1. cylindrical grasp (Cyl), 2. lumbrical grasp (Lum), 3. oblique palmar grasp (Obl), 4. intermediate power-precision grasp (IntPP), 5. two fingers pad-to-pad pinch (2-F PpP), 6. three fingers pad-to-pad pinch (3-F PpP), 7. lateral pinch (LatP) (Fig. 2(a)). The sequence of 7 grasps was the same during the experiment period. The hand grip dynamometer was used for Cyl, Lum, Obl, IntPP and the pinch dynamometer was used for 2-F PpP, 3-F PpP, LatP. At the beginning of each session, the subject would perform maximal voluntary isometric contraction to get the maximal voluntary contraction (MVC) value for the session. In each session, the subject was instructed to perform isometric ramp and hold contraction at three different intensity levels-10% MVC, 30% MVC, and 50% MVC, which corresponded to Task 1, Task 2, and Task 3. Each task comprises two same trials. In the trial lasting for 30 s, the subject was instructed to perform 5 s rest, 5 s isometric ramp up from rest to an intensity level, 10 s hold contraction, 5 s isometric ramp down to rest, and 5 s rest. The real-time force as visual feedback and trapezoidal target force curve, which the participant was instructed to trace, were displayed on the screen. The intensity level of the target force curve between 10 s and 20 s (i.e., holding contraction period) were 10% MVC, 30% MVC, 50% MVC for Task 1, Task 2, Task 3 respectively. A 30 s rest was set between trials to let the subject relax.
Experimental protocol for GRASP sub-experiment (a) and GESTURE sub-experiment (b). (a) Each day, the subject would perform 7 grasps. The sequence of 7 grasps was the same during the experiment period. In each session, the subject would perform 3 tasks of different force level. Each task contained 2 same trials lasting for 30 s: 5 s rest, 5 s isometric ramp up from rest to an intensity level, 10 s hold contraction, 5 s isometric ramp down to rest, and 5 s rest. (b) Each day, the participant would perform 5 same trials, each of them lasting for 180 s. In each trial, the participant would perform 11 gesture tasks. Each gesture task lasted for 10 s, with a 5 s rest between two neighboring tasks. There was also a 5 s rest at the beginning of the trial and 20 s rest at the end. A 180 s rest was set between trials to let the subject relax.
GESTURE sub-experiment
The whole experiment for each participant lasted for 11 days. Each day, the participant would perform 5 same trials, each of them lasting for 180 s. In each trial, the participant would perform 11 gesture tasks using their right hand: 1. hand close (HC), 2. hand open (HO), 3. wrist flexion (WF), 4. wrist extension (WE), 5. radius deviation (RD), 6. ulna deviation (UD), 7. forearm pronation (FP), 8. forearm supination (FS), 9. two fingers pad-to-pad pinch (2-F PpP), 10. three fingers pad-to-pad pinch (3-F PpP), 11. lateral pinch (LatP), which are illustrated in Fig. 2(b). Each gesture task lasted for 10 s, with a 5 s rest between two neighboring tasks. There was also a 5 s rest at the beginning of the trial and 20 s rest at the end. The ordered pictures of gestures and a time indicator were displayed on the screen to guide the subject which gesture to perform. The subject was seated with the right arm hanging naturally and not contacting the body (Fig. 1(a)(ii)). A 180 s rest was set between trials to let the subject relax.
Data processing
All HD-sEMG data of both experiments is raw data.
The force data in the GRASP experiment was normalized depending on the MVC value.
Data Records
The data presented in the article is freely accessible at Zenodo and can be downloaded from14,15. The database includes two sub-datasets: one is GRASP with 13 subjects and the other is GESTURE with 6 subjects. The data of subjects 1–10 in GRASP can be downloaded from Part I14. The data of subjects 11–13 in GRASP and all subjects in GESTURE can be downloaded from Part II15. Numeric data is in double format.
The GRASP sub-dataset contains 13 main folders for 13 subjects involved in GRASP experiment. Each folder has 462 files (11 days × 7 sessions × 3 tasks × 2 trials) in Matlab format (*.mat). Files in one day were put in a subfolder of the main folder. Each file is named in the format of SA_DayB_SessionC_TaskD_TrialE.mat, where A is the subject number (values between 1–13), B the day number (1–11), C the session number (1–7), D the Task number (1–3), E the trial number (1–2). Every file has 3 variables, namely:
data_sEMG (in mV): The HD-sEMG data in 2-D array (channels × samples). The array contains 320 rows and 61500 columns (30 s × 2048 Hz = 61440≈61500). Row 1–64 belong to electrode grid 1, row 65–128 belong to electrode gird 2, row 129–192 belong to electrode gird 3, row 193–256 belong to electrode gird 4, and row 257–320 belong to electrode gird 5. The grid number and the spatial distribution of every channel on each electrode grid can be found in Fig. 1(b–d).
data_force (dimensionless): The normalized force data in 1-D array. Columns are samples. The array length ranges from 6016 to 6048 due to slight time length variation among trials.
MVC (in kg): The maximal voluntary contraction value of each session. The values are the same for all trials in the same session on the same day.
The GESTURE sub-dataset contains 6 main folders for 6 subjects involved in GESTURE experiment, each folder has 55 files (11 days × 5 trials) in Matlab format (*.mat). Data files of one subject were also separated into different subfolders according to the experiment date. Each file is named in the format of SA_DayB_TrialC.mat, where A is the subject number (values between 1–13), B the day number (1–11), C the trial number (1–5). Every file only has one variable:
data_sEMG (in mV): The HD-sEMG data in 2-D array (channels × samples). The array contains 320 rows and 61500 columns. Row 1–64 belong to electrode grid 1, row 65–128 belong to electrode gird 2, row 129–192 belong to electrode gird 3, row 193–256 belong to electrode gird 4, and row 257–320 belong to electrode gird 5. The grid number and the spatial distribution of every channel on each electrode grid can be found in Fig. 1(b–d).
Some trials failed due to file corruption or sensor malfunction. We manually inspected all files in both sub-datasets and listed the failed files in Table 2.
Technical Validation
In addition to qualitative visual inspection of signal quality after daily experiments, the whole dataset was assessed quantitatively in an offline setting subsequently. Before the technical validation, the HD-sEMG signals were bandpass filtered through a fourth-order Butterworth filter with cut-off frequencies at 20 Hz and 500 Hz. Also, a notch filter of 50 Hz with Q factor = 35 was employed to remove powerline noise. It should be noted that the preprocessing procedure was only for technical validation and data in the dataset is raw.
Signal-to-noise Ratio
The signal-to-noise ratio (SNR) is used to quantify the amplitude increase of the signal during muscle activation compared to the background noise when the muscle is not contracting. Specifically, it can be calculated according to the following equation:
In GRASP experiment, considering the subject’s response time advance or delay, we choose 0–4.5 s and 25.5–30 s as rest time, 5.5s–24.5 s as activation time. In GESTURE experiment, we simply choose 11 gesture tasks time as activation time and the rest time in the trial as rest time. We removed the values out of the normal range (mean ± 3 standard deviation) from the data validation result. The average SNR of all trials (Fig. 3(a)) in GRASP sub-dataset (13.7372 ± 8.1597) and in GESTURE sub-dataset (16.8544 ± 5.1522) are both in accordance with the suggested SNR for some multi-finger and wrist gesture movement examples16.
(a) SNR violin plots for GRASP (upper one) and GESTURE (lower one) to show SNR values (in dB) distribution of all trials on multiple days. The x-axis represents the number of days and the y-axis represents SNR values. The green, orange, and yellow lines indicate the mean value of all SNR values of trials on the same day in Task 1 (10% MVC), Task 2 (30% MVC), and Task 3 (50% MVC). The red and black line segments in (a) indicate the median and mean value of trials on the same day. (b) CCN radar plots for GRASP (upper one) and GESTURE (lower one). Curves with different colors represent the mean value of CCN values of trials on the same day. Each axis represents different grasps (GRASP) or hand gestures (GESTURE). The nearest number to each text indicating grasps or gestures is the mean value of CCN values of trials of the grasp or gesture.
Correlation coefficient of normality
The correlation coefficient of normality (CCN) was computed to assess the resemblance between the amplitude distribution of HD-sEMG signal during activation and the normal distribution. Generally, we consider a bio-signal with quasi-normal distribution of amplitude as an uncontaminated signal. In this article, CCN value of a trial is calculated as the Pearson Correlation Coefficient between 10000 quantiles of the amplitude distribution of the whole sEMG signals (all channels) of the trial during the activation period (5–25 s) and 10000 quantiles of standardized normal distribution. The closer the value is to 1, the more similar the amplitude distribution is to the normal distribution. The mean CCN values of different grasps in GRASP sub-dataset and different gestures in GESTURE sub-dataset are shown in Fig. 3(b).
EMG-Force regression and hand gesture recognition
To verify that the data in GRASP sub-dataset allows the EMG-Force regression, we applied a linear regression model on Hudgins’s time domain features. There are two trials per task per session per day per subject. We used the first trial for training and the other trial for testing. Theoretically, there were 3003 models in total that were trained and tested (one model per task per session per day per subject; failed trials were not excluded). Initially, since all channels are not available due to poor contact between electrode and skin or electrodes overlap, we would remove these channels from the data in case that the abnormal data would affect the regression accuracy. Channels with abnormal RMS values, which meant that their RMS values were out of the range of normal values (mean ± 3 standard deviation), were considered unavailable. The union of unavailable channels of the two trials was discarded. Then, we segmented the HD-sEMG data of both trials using a sliding window of 200 ms with a 50% overlap. In total, we had 299 observations for both the training trial and the testing trial. Mean absolute value (MAV), zero crossings (ZC), slope sign changes (SSC), and waveform length (WL), were extracted for each available channel in each window17. Assume that we had C available channels (in most cases, C > 310) after channel selection, the length of the feature vector was 4 × C, which was larger than the number of samples. Hence, principal component analysis (PCA) was applied to reduce the length of the feature vector to 20018. The same sliding window was applied to force data to get the response to observation, which was computed as the mean value of the force data in each window. Optimal parameters of the linear regression model were obtained by the least square method. The model was then tested on the testing trial to validate the model performance. Three tasks in the same session (6 trials in total, S4_Day3_Session3_Task1/2/3_Trial1/2.mat) were randomly chosen to showcase the model performance (Fig. 4(a)). Coefficient of determination (\({R}^{2}\)) and normalized root-mean-square error (nRMSE) are employed as performance evaluation, whose distributions are shown in Fig. 4(b,c). The mean value of \({R}^{2}\) of all models is 0.7533 and the mean value of nRMSE of all models is 0.1359.
Panels of EMG-Force regression and hand gesture recognition. (a) Performance validation of three EMG-Force regression models. We randomly chose a session containing 3 tasks, i.e. 6 trials (S4_Day3_Session3_Task1/2/3_Trial1/2.mat). Trial 1 in each task would be used to train the model, and Trial 2 would be used to validate the model performance. Three fine red lines represent the target force the subject traced in Task1/2/3. Black lines represent the true force dynamometer recorded. Blue lines represent the force that the models predicted. (b) Boxplot for \({R}^{2}\) values of EMG-Force regression models on 7 grasps (sessions). The overall distribution plot is located at the southwest of panel (b). We then hide outliers to have a close inspection of the value distribution between nonoutlier minimum and nonoutlier maximum. (c) Boxplot for normalized root-mean-square error (nRMSE) values of EMG-Force regression models on 3 tasks of 7 grasps. Blue boxes represent nRMSE values of Task 1 (Force level is 10% MVC). Orange boxes represent nRMSE values of Task 2 (Force level is 30% MVC). Yellow boxes represent nRMSE values of Task 3 (Force level is 50% MVC). (d) The confusion matrix of the average performance of all models in hand gesture recognition. For the 5 trials in one day of one subject, 4 of them would be used to train the model and the rest one would be used for performance validation. Each model has an 11 × 11 confusion matrix. We average all confusion matrices to have the averaged matrix in (d). (e) Boxplot for within-day classification accuracies of hand gesture recognition models on multiple days. (f) Boxplot for cross-day classification accuracies of hand gesture recognition models on multiple days. For 55 trials (5 trials × 11 days) from one subject, we chose 4 trials on the first day to train the model and the rest trials for performance validation. The black curve indicates the mean value of classification accuracies of models on the same day.
To verify that the data in GESTURE sub-dataset allows hand gesture recognition, we applied a linear discrimination analysis (LDA) classification model on MAV, ZC, SSC, WL as well19. There are 5 trials per day per subject. We used four of them to train the model and the last one for testing. Theoretically, there were 66 models in total, which were trained and tested (one model per day per subject; failed trials were not excluded). A sliding window of 200 ms with a 50% overlap was used to segment the HD-sEMG data. Only the segments during activation time were kept for classification. In total, we had 1089 observations for each trial. The training set had 4356 observations while the testing set had 1089 observations. Channel selection and PCA were implemented before training the LDA model. The classification accuracy was evaluated as the ratio between the number of correctly recognized samples and tested samples. The confusion matrix indicating the average classification accuracy of all LDA models is shown in Fig. 4(d). And the classification accuracy of each day is shown in Fig. 4(e). The average accuracy of all days of all subjects is 0.9724 ± 0.0144.
Furthermore, we also test the cross-day model performance of hand gesture recognition on GESTURE sub-dataset. For each subject, Trial 1-4 in Day 1 were used to train the LDA model. Trial 5 on Day 1 and all 5 trials on other days were used to test. 6 models were trained and tested for 6 subjects. The same preprocessing procedures (channel selection and PCA) were applied before training the models. The daily validation performance of models of each subject is shown in Fig. 4(f), which suggests that the classification accuracy tends to decrease with increasing of days.
EMG decomposition
Estimating motor unit discharging patterns through HD-sEMG decomposition has demonstrated significant potential in neurophysiological research and human-machine interface applications20. We verify the two sub-datasets respectively in terms of motor unit identification. HD-sEMG decomposition was performed on each single electrode grid comprising 64 channels. HD-sEMG signals were decomposed into motor unit spike trains (MUST) based on the convolutional kernel compensation (CKC) algorithm21,22. The decoding results of 5 separate electrode arrays were combined together. Two MUSTs with the ratio of agreement23 between them larger than 0.3 are considered repeated, and the one with a greater SIL value is kept. Besides, MUST, which satisfied the following conditions, were discarded: pulse-to-noise ratio (PNR)24 <20 dB, average discharging rate >35 Hz or <5 Hz, total discharging time <critical total discharging time (5 s for GRASP trials and 3.75 s for GESTURE trials). The numbers of MUs and PNR values were used to validate the decoding result.
For GRASP sub-dataset, we randomly select 3 trials as examples (10% MVC, 30% MVC, 50% MVC) which are from the same subject, same day, and same session but different tasks (S13_Day3_Session1_ Task1/2/3_Trial1.mat) to show the decomposition result (Fig. 5(a)(i)). The multi-day distribution of numbers of MUs and PNR values are shown in Fig. 5(b)(i), (ii). The total average value of numbers of MUs and PNR values decomposed from single trial are 78.85 ± 21.92 and 27.71 ± 3.41.
(a) EMG decomposition examples. In (i), we randomly chose a session containing 3 tasks, then chose Trial 1 from each task (S13_Day3_Session1_Task1/2/3_Trial1.mat). The HD-sEMG data of these 3 trials was decomposed to extract motor unit spike trains (MUSTs). Black lines represent simultaneously recorded force data. In (ii), we randomly chose a trial as an example (S3_Day3_Trial1.mat). We segmented the data into 11 sections and decomposed each section separately. (b) Decomposition results: No. MUs and PNR values of GRASP-(i), (ii) and GESTURE-(iii), (iv). (i) Boxplot for No. MUs distribution on multiple days (GRASP). (ii) Boxplot for PNR values of MUSTs distribution on multiple days (GRASP). (iii) Boxplot for No. MUs distribution on multiple days (GESTURE). (iv) Boxplot for PNR values of MUSTs distribution on multiple days (GESTURE).
For GESTURE sub-dataset, we divided the 180 s HD-sEMG data into 11 segments, each comprising one gesture task and 2.5 s before it and 2.5 s after it. Segments were decomposed separately. We randomly choose a trial as an example (S3_Day3_Trial1.mat) to show the decomposition result (Fig. 5(a)(ii)). The multi-day distribution of numbers of MUs and PNR values can be found in Fig. 5(b)(iii),(iv). The total average value of numbers of MUs and PNR values decomposed from single segment are 59.90 ± 27.27 and 28.18 ± 3.07.
Usage Notes
The dataset presented in this paper has greatly many potential usages including but not limited to pattern recognition and EMG-Force regression, either within-day or cross-day. Researchers can compare the classification accuracy of different classifiers while choosing different features in the time domain or frequency domain. The GRASP sub-dataset contains continuous isometric force data simultaneously recorded with HD-sEMG data, thus the proportional control method can also be studied using the data. In addition, since our dataset comprises 320-channel high-density surface electromyography signals covering almost the whole forearm region, readers may obtain the motor unit action potential trains (MUAPTs) via decomposition of HD-sEMG and investigate the neuromuscular physiological state of the right forearm performing different gestures over multiple days.
We encourage any usage which is of contribution to the advancing research on neuro-physiology and muscle synergistic activities as well as the development of robust HMI and intuitively controlled prostheses based on sEMG. It is important to note that No. 6 subject in GESTURE missed the experiment on Day 7, thus he extended the experiments till Day 12 for compensation. However, the file names of subject No. 6 are still D1…D11 for uniformity.
Code availability
We provide some functions to process the raw data in the dataset, such as filtering and available channels selection. The functions are in the folder named MyFunctions, together with a code script as an instrumental example15.
References
Hermens, H. J., Freriks, B., Disselhorst-Klug, C. & Rau, G. Development of recommendations for SEMG sensors and sensor placement procedures. Journal of electromyography and kinesiology: official journal of the International Society of Electrophysiological Kinesiology 10(5), 361–374, https://doi.org/10.1016/s1050-6411(00)00027-4 (2000).
Jiang, N. et al. Bio-robotics research for non-invasive myoelectric neural interfaces for upper-limb prosthetic control: a 10-year perspective review. National science review 10(5), nwad048, https://doi.org/10.1093/nsr/nwad048 (2023).
Zheng, M., Crouch, M. S. & Eggleston, M. S. Surface Electromyography as a Natural Human–Machine Interface: A Review. IEEE Sensors Journal 22, 9198–9214 (2021).
Balbinot, G. et al. The use of surface EMG in neurorehabilitation following traumatic spinal cord injury: A scoping review. Clinical neurophysiology: official journal of the International Federation of Clinical Neurophysiology 138, 61–73, https://doi.org/10.1016/j.clinph.2022.02.028 (2022).
Howard, R. M., Conway, R. & Harrison, A. J. Muscle activity in sprinting: a review. Sports biomechanics 17(1), 1–17, https://doi.org/10.1080/14763141.2016.1252790 (2018).
Li, W., Shi, P. & Yu, H. Gesture Recognition Using Surface Electromyography and Deep Learning for Prostheses Hand: State-of-the-Art, Challenges, and Future. Frontiers in neuroscience 15, 621885, https://doi.org/10.3389/fnins.2021.621885 (2021).
Campbell, E., Phinyomark, A. & Scheme, E. Current Trends and Confounding Factors in Myoelectric Control: Limb Position and Contraction Intensity. Sensors (Basel, Switzerland) 20(6), 1613, https://doi.org/10.3390/s20061613 (2020).
Song, T. et al. Review of sEMG for Robot Control: Techniques and Applications. Applied Sciences. 13(17), 9546, https://doi.org/10.3390/app13179546 (2023).
Zhu, X., Liu, J., Zhang, D., Sheng, X. & Jiang, N. Cascaded Adaptation Framework for Fast Calibration of Myoelectric Control. IEEE transactions on neural systems and rehabilitation engineering: a publication of the IEEE Engineering in Medicine and Biology Society 25(3), 254–264, https://doi.org/10.1109/TNSRE.2016.2562180 (2017).
Matran-Fernandez, A., Rodríguez Martínez, I. J., Poli, R., Cipriani, C. & Citi, L. SEEDS, simultaneous recordings of high-density EMG and finger joint angles during multiple hand movements. Scientific data 6(1), 186, https://doi.org/10.1038/s41597-019-0200-9 (2019).
Pradhan, A., He, J. & Jiang, N. Multi-day dataset of forearm and wrist electromyogram for hand gesture recognition and biometrics. Scientific data 9(1), 733, https://doi.org/10.1038/s41597-022-01836-y (2022).
Du, Y., Jin, W., Wei, W., Hu, Y. & Geng, W. Surface EMG-Based Inter-Session Gesture Recognition Enhanced by Deep Domain Adaptation. Sensors (Basel, Switzerland) 17(3), 458, https://doi.org/10.3390/s17030458 (2017).
Jiang, X. et al. Open Access Dataset, Toolbox and Benchmark Processing Results of High-Density Surface Electromyogram Recordings. IEEE transactions on neural systems and rehabilitation engineering: a publication of the IEEE Engineering in Medicine and Biology Society 29, 1035–1046, https://doi.org/10.1109/TNSRE.2021.308255 (2021).
Yang, S., Li, D., Chen, C. & Zhu, X. Dataset: CEMHSEY (ConsecutivE Multi-day High-density Surface ElectromyographY), Part I [Data set]. Zenodo https://doi.org/10.5281/zenodo.14224328 (2025).
Yang, S., Li, D., Chen, C. & Zhu, X. Dataset: CEMHSEY (ConsecutivE Multi-day High-density Surface ElectromyographY), Part II [Data set]. Zenodo https://doi.org/10.5281/zenodo.14272463 (2025).
Botros, F. S., Phinyomark, A. & Scheme, E. J. Electromyography-Based Gesture Recognition: Is It Time to Change Focus From the Forearm to the Wrist? IEEE Transactions on Industrial Informatics 18, 174–184 (2022).
Phinyomark, A., Phukpattaranont, P. & Limsakul, C. Feature reduction and selection for EMG signal classification. Expert Syst. Appl. 39, 7420–7431 (2012).
Zhang, D., Xiong, A., Zhao, X., & Han, J. PCA and LDA for EMG-based control of bionic mechanical hand. 2012 IEEE International Conference on Information and Automation, 960-965 (2012).
Fisher, R. A. The use of multiple measurements in taxonomic problems. Annals of Human Genetics 7, 179–188 (1936).
Chen, C., Yu, Y., Sheng, X. & Zhu, X. Non-Invasive Analysis of Motor Unit Activation During Simultaneous and Continuous Wrist Movements. IEEE journal of biomedical and health informatics 26(5), 2106–2115, https://doi.org/10.1109/JBHI.2021.3135575 (2022).
Holobar, A. & Zazula, D. Multichannel Blind Source Separation Using Convolution Kernel Compensation. IEEE Transactions on Signal Processing 55, 4487–4496 (2007).
Holobar, A. & Zazula, D. Gradient Convolution Kernel Compensation Applied to Surface Electromyograms. International Conference on Agents (2007).
Clarke, A. K. et al. Deep Learning for Robust Decomposition of High-Density Surface EMG Signals. IEEE transactions on bio-medical engineering 68(2), 526–534, https://doi.org/10.1109/TBME.2020.3006508 (2021).
Holobar, A., Minetto, M. A. & Farina, D. Accurate identification of motor unit discharge patterns from high-density surface EMG and validation with a novel signal-based performance metric. Journal of neural engineering 11(1), 016008, https://doi.org/10.1088/1741-2560/11/1/016008 (2014).
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant Nos. 52205024, 52227808).
Author information
Authors and Affiliations
Contributions
C.C. conceived and designed the experiment. Y.S. conducted the experiment, analyzed the data, and wrote the manuscript. C.C. and L.D. helped conduct the experiment, analyzed the results, and optimized the data presentation. All authors revised the manuscript and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, S., Chen, C., Li, D. et al. A Consecutive Multi-Day High-Density Surface Electromyography Dataset Comprising 7 Grasps and 11 Gestures. Sci Data 12, 1420 (2025). https://doi.org/10.1038/s41597-025-05733-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05733-y