Abstract
Prototheca, one of the foremost causative agents responsible for opportunistic infections in both humans and animals. Despite the availability of several Prototheca genomes, the genomes of Prototheca bovis and Prototheca ciferrii have remained unsequenced. In this study, we report two high-quality genome assemblies for P. bovis strain SH08 and P. ciferrii strain SH13, representing two distinct species of the genus Prototheca. The final assembled genome sizes were 30.6 Mb for P. bovis SH08 and 32.7 Mb for P. ciferrii SH13, with contig N50 values of 1.19 Mb and 1.78 Mb, respectively. The repetitive sequences were identified in 14.79% and 14.24% of the assembled genomes, respectively. A total of 5,141 protein-coding genes were predicted for P. bovis SH08, while P. ciferrii SH13 contained 4,986 protein-coding genes. Functional annotation identified 97.28% and 99.16% of the genes in the genomes, respectively. These two novel high-quality genome assemblies represent valuable resources for advancing research in evolutionary biology, comparative genomics, pathogenicity assessment, diagnostic strategies, and therapeutic interventions in protothecosis.
Similar content being viewed by others
Background & Summary
Prototheca is a genus of achlorophyllous green microalgae classified within the phylum Chlorophyta, class Trebouxiophyceae, and family Chlorellaceae. It exhibits wide distribution across diverse environments worldwide1. Prototheca has been identified as a pathogen capable of infecting humans (primarily caused by Prototheca wickerhamii), cattle and dog (mainly caused by Prototheca bovis and Prototheca ciferrii), and some other reported species caused by Prototheca cutis, Prototheca blaschkeae and Prototheca miyajii1,2,3. The Prototheca infection, also known as Protothecosis, has been documented in several hundred cases4. Over the past two decades, there has been a significant increase in the prevalence of Prototheca as an emerging pathogen5. However, several knowledge gaps persist regarding their biological characteristics, particularly concerning their pathogenicity. In particular, the limited genomic and molecular research on Prototheca impedes the development of robust diagnostic tools and a comprehensive understanding of its pathogenesis. To enhance public awareness of protothecosis and improve its identification and diagnosis, we have initiated and established the Protothecosis Science Popularization and Monitoring Consortium (PSPMC) and China Prototheca Working Group (CPWG) in collaboration with multiple organizations.
Due to the rapid development and cost reduction of next-generation long-read sequencing technologies, genome sequencing has become more cost-effective6,7. For the Prototheca genus, several genomes have been assembled, including P. wickerhamii strain ATTC165294, P. wickerhamii strain S1 and S9318, P. wickerhamii strain InPu-22_FZ9 and two strains of Prototheca zopfii Pz20 and Pz2310. In addition to genome reference resources, multi-omics approaches such as transcriptomics, proteomics, and metabolomics have been employed to further elucidate the pathogenic mechanisms of Prototheca11. The diagnosis of Prototheca infection is generally determined through blood, body fluid, or tissue culture12. However, it poses a significant challenge, particularly in cases where protothecosis is not suspected clinically. The diagnosis cannot be classified as different Prototheca without DNA sequencing. The genomic data of different Prototheca species not only enhances diagnostic precision but also provides valuable insights for foundational research. A total of eighteen Prototheca species have been reported, with fifteen being documented in 201913, and three newly identified species, Prototheca fontanea, Prototheca lentecrescens, and Prototheca vistulensis were later characterized14. However, all the taxonomic samples utilized in the study were obtained from Poland based on the cytochrome B(CYTB)-based PCR-restriction fragment length polymorphism (RFLP). P. bovis and P. ciferrii, previously classified as P. zopfii genotype 1 and genotype 2 respectively5,15, are closely associated with dairy herd environments and remain under investigation. In this study, samples of the two species were collected from China. The species P. bovis has been identified as the most pathogenic among dairy cattle16, while P. ciferrii has been reported that it can cause the infections in dogs inducing a more aggressive disease course17,18. A high-quality genome serves as the genetic foundation for molecular research and gene diagnostics. Recently, high-throughput long-read sequencing, particularly PacBio HiFi sequencing, has enabled the generation of the highest-quality genomes to date19. The genome assembly algorithms have revolutionized the field by significantly improving computational efficiency and reducing costs19. These two high-quality genome resources hold significant value for taxonomy identification, evolutionary studies, comprehensive understanding, and even the diagnosis of Protothecosis.
Methods
The process of isolating and obtaining the Prototheca strains was approved by the Medical Ethics Committee of Shanghai East Hospital, Tongji University (Approval No. 2024YS-274). An overview of the experimental and bioinformatic workflow used in this study is depicted in Fig. 1. Briefly, the process encompassed sample collection, DNA extraction, multi-platform sequencing, genome assembly, annotation, and comprehensive quality assessment.
Sample collection and extraction
The strain of Prototheca bovis SH08 was obtained from fresh cow milk sample in Shanghai, China. Prototheca ciferrii SH13 was obtained from skin tissue samples of human patients in Shanghai, China. The two strains were classified as P. bovis and P. ciferrii based on the previous protocol and analysis of the CYTB gene14. The samples of P. bovis SH08 and P. ciferrii SH13 can be available at the biobank of Shanghai East Hospital, Tongji University. The two strains were cultivated in a bacteriological incubator on Sabouraud dextrose agar medium and incubated at 35 °C for 4 days, followed by subsequent harvesting for DNA extraction. The high molecular weight (HMW) genomic DNA of the P. bovis SH08 and P. ciferrii SH13 were extracted using the CTAB method, following a previously reported protocol20.
Short library construction, sequencing, and genome survey
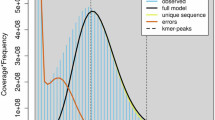
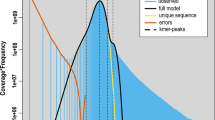
The short reads library was generated for P. bovis SH08. The library with an insert size of 300–500 bp was subjected to Covaris E220 System for fragmentation and prepared using the MGIEasy Universal DNA Library Prep Set (MGI-Tech). The PE 150 reads were sequenced on the next generation sequencing MGISEQ-2000 platform, generating approximately 5.5 gigabases (Gb) of sequencing data (Table 1), following identical filter parameters as our previous study10. The 17-mer and 19-mer frequency were calculated for P. bovis SH08 using jellyfish21. The estimated genome sizes are approximately 29.06 Mb and 30.1 Mb, with a total of 4,621,095,510 and 4,551,930,609 k-mers, and peak depths of 159 and 151, respectively, as calculated by KmerFreq v5.021.
PacBio Library construction, sequencing, and genome assembly
For PacBio HiFi sequencing, two libraries with an approximate insert size of 20 kb were generated for P. bovis SH08 and P. ciferrii SH13 using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, USA). A total of approximately 1.99 Gb (119,083 reads) and 4.17 Gb (242,272 reads) clean data were generated using PacBio SequelII SMRT cell with HiFi model, respectively (Table 1). The average length of CCS reads was 16,720 bp and 17,214 bp for P. bovis SH08 and P. ciferrii SH13, respectively (Table 1). Using the HiFi long reads, the nuclear genomes of the P. bovis SH08 and P. ciferrii SH13 were assembled using hifiasm v0.7 with default parameters22. The assembled genome of P. bovis SH08 is 30.6 Mb, with a contig N50 of 1.19 Mb and a total number of 53 contigs. Similarly, the assembled genome of P. ciferrii SH13 is 32.7 Mb, with a contig N50 of 1.78 Mb and a total number of 96 contigs (Fig. 2 and Table 2). The maximum length of assembled genome of P. bovis SH08 and P. ciferrii SH13 is 2,899,485 bp and 2,975,726 bp, respectively (Table 1). The GC content of P. bovis SH08 and P. ciferrii SH13 is 73.6% and 67.8%, respectively (Table 1).
Genome annotation
The repeat annotation was conducted by employing TRF (4.9) with default parameters for the identification of Tandem repeats23. Then, the homologous sequences of the P. bovis SH08 and P. ciferrii SH13 genomes were identified using the software RepeatProteinMask (v 4.0.7)24 and RepeatMasker (open-4.0.9)25 based on the Repbase library (http://www.girinst.org/repbase)26. The databases of two strains repetitive sequence were generated using RepeatModeler open-1.0.1140 and LTR_FINDER_parallel 1.0.741, followed identified using the ab initio method with RepeatMasker (open-4.0.9)25. Finally, 14.79% and 14.24% of assembled P. bovis SH08 and P. ciferrii SH13 genomes were classified as repetitive sequences (Table 2 and Table 3). After repeat identification, three different algorithms, namely homology-based annotation, ab initio prediction and RNA-Seq data-based annotation were employed for gene prediction. In homology-based annotation, a total of 10 published homology protein sequences from Coccomyxa subellipsoidea, Chlorella variabilis, Chlorella desiccate, Chlorella sorokiniana, Auxenochlorella protothecoides, three strains of Prototheca wickerhamii ATTC16529, S1 and S931, and two strains of Prototheca zopfii Pz20 and Pz23 were integrated with the MAKER2 pipeline27. In the ab initio prediction, the P. bovis SH08 and P. ciferrii SH13 genome sequences, which were masked for repeated elements, were employed to identify coding regions of genes using AUGUSTUS v3.2.328 and SNAP29. For RNA-based prediction, the relative transcriptome of P.zopfii (7.5 Gb) from the published study30. The RNA reads were mapped to the genomes of two strains of P. bovis SH08 and P. ciferrii SH13 genomes using hista2.2.131, followed by transcript assembly using stringtie2.1.632. The gene sets were obtained by integrating the three strategies using Maker2 (2.31.10)27. Finally, a total of 5,141 and 4,986 protein-coding genes were obtained respectively for P. bovis SH08 and P. ciferrii SH13 genomes (Table 2). The total count of gene models exhibiting an Annotation Edit Distance (AED) score ≤ 0.5 reached 4,957 (96.4%) for P. bovis SH08 and 4,805 (96.4%) for P. ciferrii SH13, indicating robust support from the available evidence.
Functional annotation
The gene set of the P. bovis SH08 and P. ciferrii SH13 genomes was functionally annotated based on the seven databases including NR version 2023-04-01 (NCBI nonredundant protein), KEGG version 105.0, 2023-01-01 (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/), KOG version 2023-03-0147, TrEMBL version 2023-03-01 (http://www.uniprot.org), Swiss-Prot version 2023-03-01 (http://www.gpmaw.com/html/swiss-prot.html), InterPro 93.0 and GO Ontology (GO) version 2023-04-01. The functional annotation for P. bovis SH08 and P. ciferrii SH13 genomes accounted for 97.28% and 99.16% of the annotated genes, respectively (Table 4). A total of 4,344 (84.50%) and 4,175 (83.73%) genes can be functional annotation in KEGG database (Fig. 3). The primary metabolic pathway identified is Carbohydrate metabolism (387), with P. bovis SH08 and P. ciferrii SH13 exhibiting significant involvement in Carbohydrate metabolism (365) as shown in Fig. 3. A total of 3,577 and 3,529 genes can be annotated in all seven databases for P. bovis SH08 and P. ciferrii SH13, respectively (Fig. 4).
Ethics statement
The Ethics Committee of Shanghai East Hospital, Tongji University approved the study protocol and the sharing of de-identified genomic data. Informed consent was obtained from all participants. The ethical approval for this study includes the isolation of Prototheca from the milk of the cows and sample of the patients, and the consent of the dairy farm manager was obtained when collecting the milk samples.
Technical Validation
Evaluation of the genome assembly
The quality assessment of the two strain genomes was conducted by aligning the HiFi reads to a high-quality assembled genome using Minimap2, resulting in a mapping rate and coverage of 96.91% and 98.57% for P. bovis SH08, and a mapping rate and coverage of 96.35% and 96.71% for P. ciferrii SH13, respectively. The mean depth of mapping coverage is 60.15 × and 114.37 × for P. bovis SH08 and P. ciferrii SH13, respectively. The distribution of sequencing coverage depth and accumulative coverage showed that the genome assembly is high quality (Fig. 5) with 10 kb window. The accuracy of the final assembly of the two strains was assessed using Merqury with a 19-mer, yielding a quality value (QV) score of 56.4816, indicating an accuracy rate of 99.99977% for SH08 and a QV score of 57.2924, reflecting an accuracy rate of 99.99981% for SH13. The genome sizes of P. bovis SH08 and P. ciferrii SH13 are comparable to those of P. zopfii Pz20 and Pz23 (~31 Mb), while the contig N50 values for P. bovis SH08 and P. ciferrii SH13 exceed 1 Mb, indicating exceptionally high-quality genome assemblies. The analysis of GC content revealed that the genomes of P. bovis SH08 and P. ciferrii SH13 exhibit a GC content of 73.6% and 67.8% (Fig. 6), respectively, indicating the absence of exogenous species contamination in both species. To mitigate potential contamination, the HiFi assembled genomes were aligned against the GenBank nucleotide (nt) database, and the available Prototheca organelle Refseq genome was retrieved from NCBI. The contigs exhibited no bacterial contamination, and only a limited number of organelle sequences were identified and subsequently excluded from the nuclear genomes. The completeness of the P. bovis SH08 and P. ciferrii SH13 genome sequences were evaluated using BUSCO, based on the “chlorophyta_odb10” database. The analysis revealed that 76.7% and 78.2% of the 1519 conserved chlorophyta genes were identified as complete (Table 5), which is comparable to other Prototheca species.
Evaluation of the gene set
Based on the comparison of gene features, the distribution of gene(mRNA) length and coding sequence (CDS) length exhibited remarkable similarity among the eight green algae (Fig. 7a, c). The P. bovis SH08 and P. ciferrii SH13 exhibited a similar pattern to P. zopfii Pz20 and Pz23 in terms of exon length, intron length, and exon number, which is consistent with their comparable genome size and relatedness (Fig. 7b, d, e). Furthermore, the gene features indicated that the gene annotation is of high quality when compared to published data. The completeness of the P. bovis SH08 and P. ciferrii SH13 gene sets were evaluated using BUSCO, based on the “chlorophyta_odb10” database. The analysis revealed that 76.7% and 77.7% of the 1519 conserved chlorophyta genes were identified as complete (Fig. 8), indicating high-quality gene annotation comparable to other Prototheca species (77.3% and 77.6% in P. zopfii Pz20 and Pz23)10.
Genome sequences comparison
To confirm the genome quality, the published P. bovis SAG 2021 and P. zoffii Pz20 genomes were downloaded. The published P. bovis SAG 2021 genome assembly comprised 24,744,895 bp and consisted of 4,555 contigs15. The previously fragmented P. bovis SAG 2021 genome was scaffolded and patched using the RagTag software33, based on our newly assembled reference P. bovis SH08 genome. A total of 3,815 contigs were placed, with a total length of 21,558,679 bp (87.1% of assembled genome size). Then, two genomes were compared using MUMmer4 with nucmer model (c 2000 -l 400). The mummer analysis of P. bovis genomes revealed clearly and high similarity (>97%), and suggested genome assembly was accurate (Fig. 9a). The newly P. ciferrii SH13 genome was compared with previous published P. zoffii Pz20 genome also using MUMmer4 with nucmer model (c 2000 -l 400). The mummer analysis of genomes revealed nearly one contig to one contig clearly and high similarity (>96%) and suggested genome assembly was correct (Fig. 9b).
Data Records
The P. bovis SH08 and P. ciferrii SH13 genome sequences and annotations were deposited in Figshare.The dataset had been deposited at PRJNA1184973 in the Sequence Read Archive (SRA)34. DNBSeq short-read data of P. bovis SH08 was deposited in the SRA at SRR31320872. HiFi long-read data of P. bovis SH08 and P. ciferrii SH13 were deposited in the SRA at SRR31320874 and SRR31320873. The genome assembly of P. bovis SH08 and P. ciferrii SH13 had been deposited at GenBank under accession JBJGBU00000000035 and JBJGBT00000000036. The P. bovis SH08 and P. ciferrii SH13 genome sequences and annotations were deposited in Figshare37.
Data availability
The dataset of DNB-Seq short-read, HiFi long-read had been deposited in the Sequence Read Archive (SRA) under project number PRJNA118497334. The link of sequencing data was provided below:
DNBSeq short-read data of P. bovis SH08 was deposited in the SRA at SRR31320872.
HiFi long-read data of P. bovis SH08 was deposited in the SRA at SRR31320874.
HiFi long-read data of P. ciferrii SH13 was deposited in the SRA at SRR31320873.
The genome assembly of P. bovis SH08 had been deposited at GenBank under accession JBJGBU00000000035. The genome assembly of P. ciferrii SH13 had been deposited at GenBank under accession JBJGBT00000000036.
The P. bovis SH08 and P. ciferrii SH13 genome sequences and annotations were deposited in Figshare37.
Code availability
This study did not involve the development of any specific code. The data analyses were conducted in accordance with the protocols outlined in the Methods section.
References
Leimann, B. C., Monteiro, P. C., Lazéra, M., Candanoza, E. R. & Wanke, B. Protothecosis. Medical mycology 42, 95–106, https://doi.org/10.1080/13695780310001653653 (2004).
Kwiecinski, J. Biofilm formation by pathogenic Prototheca algae. Letters in applied microbiology 61, 511–517, https://doi.org/10.1111/lam.12497 (2015).
Todd, J. R. et al. Medical phycology 2017. Medical mycology 56, S188–s204, https://doi.org/10.1093/mmy/myx162 (2018).
Bakula, Z. et al. A first insight into the genome of Prototheca wickerhamii, a major causative agent of human protothecosis. BMC Genomics 22, 168, https://doi.org/10.1186/s12864-021-07491-8 (2021).
Libisch, B. et al. Prototheca Infections and Ecology from a One Health Perspective. Microorganisms 10, https://doi.org/10.3390/microorganisms10050938 (2022).
Espinosa, E., Bautista, R., Larrosa, R. & Plata, O. Advancements in long-read genome sequencing technologies and algorithms. Genomics 116, 110842, https://doi.org/10.1016/j.ygeno.2024.110842 (2024).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics 17, 333–351, https://doi.org/10.1038/nrg.2016.49 (2016).
Guo, J. et al. Genome Sequences of Two Strains of Prototheca wickerhamii Provide Insight Into the Protothecosis Evolution. Front Cell Infect Microbiol 12, 797017, https://doi.org/10.3389/fcimb.2022.797017 (2022).
Li, J. et al. Complete genome identified of clinical isolate Prototheca. Journal of Medical Microbiology 73, https://doi.org/10.1099/jmm.0.001914 (2024).
Jian, J. et al. Two high-quality Prototheca zopfii genomes provide new insights into their evolution as obligate algal heterotrophs and their pathogenicity. Microbiology spectrum 12, e04148–04123, https://doi.org/10.1128/spectrum.04148-23 (2024).
Guo, J. et al. Integration of transcriptomics, proteomics, and metabolomics data for the detection of the human pathogenic Prototheca wickerhamii from a One Health perspective. Front Cell Infect Microbiol 13, 1152198, https://doi.org/10.3389/fcimb.2023.1152198 (2023).
Cullen, G. D., Yetmar, Z. A., Fida, M. & Abu Saleh, O. M. Prototheca Infection: A Descriptive Study. Open forum infectious diseases 10, ofad294, https://doi.org/10.1093/ofid/ofad294 (2023).
Jagielski, T. et al. The genus Prototheca (Trebouxiophyceae, Chlorophyta) revisited: Implications from molecular taxonomic studies. Algal Research 43, 101639, https://doi.org/10.1016/j.algal.2019.101639 (2019).
Jagielski, T. et al. Occurrence of Prototheca Microalgae in Aquatic Ecosystems with a Description of Three New Species, Prototheca fontanea, Prototheca lentecrescens, and Prototheca vistulensis. Applied and environmental microbiology 88, e0109222, https://doi.org/10.1128/aem.01092-22 (2022).
Severgnini, M. et al. Genome sequencing of Prototheca zopfii genotypes 1 and 2 provides evidence of a severe reduction in organellar genomes. Scientific Reports 8, 14637, https://doi.org/10.1038/s41598-018-32992-0 (2018).
Huilca-Ibarra, M. P. et al. High Prevalence of Prototheca bovis Infection in Dairy Cattle with Chronic Mastitis in Ecuador. Veterinary sciences 9, https://doi.org/10.3390/vetsci9120659 (2022).
Stenner, V. J. et al. Protothecosis in 17 Australian dogs and a review of the canine literature. Medical mycology 45, 249–266, https://doi.org/10.1080/13693780601187158 (2007).
Silveira, C. S. et al. A Case of Prototheca zopfii Genotype 1 Infection in a Dog (Canis lupus familiaris). Mycopathologia 183, 853–858, https://doi.org/10.1007/s11046-018-0274-5 (2018).
Jian, J., Gao, Q., Cheng, J. & Yin, Y. Genome Assembly Algorithms. IntechOpen. https://doi.org/10.5772/intechopen.1011544 (2025).
Jagielski, T. et al. An optimized method for high quality DNA extraction from microalga Prototheca wickerhamii for genome sequencing. Plant Methods 13, 77, https://doi.org/10.1186/s13007-017-0228-9 (2017).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770, https://doi.org/10.1093/bioinformatics/btr011 (2011).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18, 170–175, https://doi.org/10.1038/s41592-020-01056-5 (2021).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580, https://doi.org/10.1093/nar/27.2.573 (1999).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11, https://doi.org/10.1186/s13100-015-0041-9 (2015).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl 1), i351–358, https://doi.org/10.1093/bioinformatics/bti1018 (2005).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research 110, 462–467, https://doi.org/10.1159/000084979 (2005).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491, https://doi.org/10.1186/1471-2105-12-491 (2011).
Stanke, M., Schöffmann, O., Morgenstern, B. & Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62, https://doi.org/10.1186/1471-2105-7-62 (2006).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59, https://doi.org/10.1186/1471-2105-5-59 (2004).
Jian, J. et al. Two high-quality Prototheca zopfii genomes provide new insights into their evolution as obligate algal heterotrophs and their pathogenicity. Microbiology spectrum 12, e0414823, https://doi.org/10.1128/spectrum.04148-23 (2024).
Yang, Z. et al. Convergent horizontal gene transfer and cross-talk of mobile nucleic acids in parasitic plants. Nature Plants 5, 991–1001, https://doi.org/10.1038/s41477-019-0458-0 (2019).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 20, 278, https://doi.org/10.1186/s13059-019-1910-1 (2019).
Alonge, M. et al. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biology 23, 258, https://doi.org/10.1186/s13059-022-02823-7 (2022).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP544791 (2025).
Guo, J. Prototheca bovis strain SH08 and Prototheca ciferrii strain SH13 Genome sequencing and assembly. GenBank https://identifiers.org/ncbi/insdc:JBJGBU000000000 (2025).
Guo, J. Prototheca bovis strain SH08 and Prototheca ciferrii strain SH13 Genome sequencing and assembly. GenBank https://identifiers.org/ncbi/insdc:JBJGBT000000000 (2025).
Guo, J. Two high-quality genomes of Prototheca bovis strain SH08 and Prototheca ciferrii strain SH13. figshare. Dataset https://doi.org/10.6084/m9.figshare.29379134.v2 (2025).
Acknowledgements
This work is financially supported by the Shanghai Pudong New Area Health Commission General Project (Grant No. PW2024A-02) and the Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No.W2411075). We are very grateful to Professor Jianbo Jian from Shantou University, who has been our close partner for a long time and has provided us with great assistance in our research.
Author information
Authors and Affiliations
Contributions
C. Jiang, W. Wu conceived the study. J. Guo, B. Luo, H. Zhu collected the samples, conducted experiments, Y. Ming, Y. Peng performed bioinformatics analysis. J. Guo, Y. Ming and Y. Xue wrote the manuscript. W. Wu provided suggestion and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, J., Ming, Y., Chen, J. et al. Two high-quality genomes of Prototheca bovis strain SH08 and Prototheca ciferrii strain SH13. Sci Data 13, 23 (2026). https://doi.org/10.1038/s41597-025-06337-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-06337-2