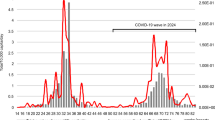
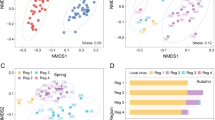
Abstract
Wastewater monitoring for pathogen detection has greatly advanced over the course of the COVID-19 pandemic. While most wastewater surveillance programs only target specific pathogens using qPCR or amplicon sequencing, untargeted wastewater metatranscriptomic sequencing (W-MTS) offers broader detection capabilities. However, there is a lack of data allowing the comparison of W-MTS with more established detection methods. Here we present a dataset consisting of 13.1 terabases (43B read pairs) of untargeted Illumina W-MTS data, generated from 20 wastewater samples, with 1.4B to 2.8B 150 bp read pairs per sample. Wastewater samples were collected between December 2023 and April 2024 at the Hyperion Water Reclamation Plant (HWRP), Los Angeles, USA, serving a population of approximately 4 million residents. The resulting dataset, one of the largest W-MTS collections to date, contains bacterial, archaeal, eukaryotic, and viral taxa—including human-infecting viruses—and many sequences of unknown origin. Uploaded to the NCBI Sequence Read Archive, we expect this data to spur additional research into the viability of pathogen-agnostic wastewater epidemiology and pathogen early detection.
Similar content being viewed by others
Data availability
All raw metatranscriptomic sequencing data generated in this study are deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA119800130. The dataset contains paired-end FASTQ files (2 × 150 bp) for each wastewater sample, organized into BioSamples SAMN45825509–SAMN45825528 and corresponding SRA Experiments SRX27073143–SRX2707316231. Metadata describing sample collection dates, sample type, and sequencing platform are included in each BioSample record. BioSample metadata follows the Genomic Standards Consortium’s “MIMS Environmental/Metagenome” metadata standard. The analysis results can be accessed in the following figshare repository: https://doi.org/10.6084/m9.figshare.28454990.v132.
Code availability
Sequencing data quality and taxonomic composition was assessed using a comprehensive computational pipeline, available under https://github.com/naobservatory/mgs-workflow/tree/2.5.1. The analysis results can be accessed in the following figshare repository: https://doi.org/10.6084/m9.figshare.28454990.v132. Code for figures and tables can be accessed under https://github.com/naobservatory/w-mgs-data-paper.
References
Dattani, S. & Roser, M. What were the death tolls from pandemics in history? Our World in Data (2023).
Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018).
Li, Y. et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet 399, 2047–2064 (2022).
Wolfe, M. K. et al. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in northern California shows strong association with COVID-19 incidence. mSystems 6, e0082921 (2021).
Brinkman, N. E., Fout, G. S. & Keely, S. P. Retrospective Surveillance of Wastewater To Examine Seasonal Dynamics of Enterovirus Infections. mSphere 2, https://doi.org/10.1128/msphere.00099-17 (2017).
McCall, C., Wu, H., Miyani, B. & Xagoraraki, I. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 184, 116160 (2020).
Randazza-Pade, J. Pathogen Biomarkers in Wastewater, Stool and Urine. https://biobot.io/pathogen-biomarkers-in-wastewater-stool-and-urine-nearly-endless-opportunities-for-the-future-of-wbe/.
Diemert, S. & Yan, T. Clinically unreported salmonellosis outbreak detected via comparative genomic analysis of municipal wastewater Salmonella isolates. Appl. Environ. Microbiol. 85 (2019).
Zhao, Y. et al. Strain-level multidrug-resistant pathogenic bacteria in urban wastewater treatment plants: Transmission, source tracking and evolution. Water Res. 267, 122538 (2024).
Pronyk, P. M. et al. Advancing pathogen genomics in resource-limited settings. Cell Genom 3, 100443 (2023).
Keshaviah, A. et al. Wastewater monitoring can anchor global disease surveillance systems. The Lancet Global Health 11, e976–e981 (2023).
Adalja, A. A., Watson, M., Toner, E. S., Cicero, A. & Inglesby, T. V. The Characteristics of Pandemic Pathogens. (2018).
Crits-Christoph, A. et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. MBio 12 (2021).
Spurbeck, R. R., Catlin, L. A., Mukherjee, C., Smith, A. K. & Minard-Smith, A. Analysis of metatranscriptomic methods to enable wastewater-based biosurveillance of all infectious diseases. Frontiers in Public Health 11 (2023).
Rothman, J. A. et al. RNA Viromics of Southern California Wastewater and Detection of SARS-CoV-2 Single-Nucleotide Variants. Appl. Environ. Microbiol. 87, e01448–21 (2021).
Child, H. T. et al. Comparison of metagenomic and targeted methods for sequencing human pathogenic viruses from wastewater. MBio 14, e0146823 (2023).
Bengtsson-Palme, J. et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 572, 697–712 (2016).
Brinch, C. et al. Long-Term Temporal Stability of the Resistome in Sewage from Copenhagen. mSystems 5, e00841–20 (2020).
Langenfeld, K. et al. Development of a quantitative metagenomic approach to establish quantitative limits and its application to viruses. bioRxiv https://doi.org/10.1101/2022.07.08.499345 (2022).
Maritz, J. M., Ten Eyck, T. A., Elizabeth Alter, S. & Carlton, J. M. Patterns of protist diversity associated with raw sewage in New York City. ISME J. 13, 2750–2763 (2019).
Munk, P. et al. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 13, 7251 (2022).
Ng, C. et al. Metagenomic and Resistome Analysis of a Full-Scale Municipal Wastewater Treatment Plant in Singapore Containing Membrane Bioreactors. Front. Microbiol. 10 (2019).
Rothman, J. A. et al. Longitudinal metatranscriptomic sequencing of Southern California wastewater representing 16 million people from August 2020-21 reveals widespread transcription of antibiotic resistance genes. Water Res. 229, 119421 (2023).
Wyler, E. et al. Pathogen dynamics and discovery of novel viruses and enzymes by deep nucleic acid sequencing of wastewater. Environ. Int. 190, 108875 (2024).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Lu, J. et al. Metagenome analysis using the Kraken software suite. Nat. Protoc. 1–25 (2022).
Index zone by BenLangmead. https://benlangmead.github.io/aws-indexes/k2.
BBMap. SourceForge https://sourceforge.net/projects/bbmap/ (2022).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA1198001 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP551368 (2024).
Grimm, S. Output of https://github.com/naobservatory/mgs-workflow/tree/2.5.0, used for Deep wastewater metatranscriptomic sequencing data, Los Angeles, USA, 2023-2024. figshare https://doi.org/10.6084/M9.FIGSHARE.28454990.V1 (2025).
Acknowledgements
S.L.G., J.T.K., W.J.B., K.L.W., and J.A.R. were funded for this research project by gifts from Open Philanthropy (to SecureBio). J.A.R was supported by an allocation (#BIO240238) from the National Science Foundation Advanced Cyberinfrastructure Coordination Ecosystem: Services and Support (ACCESS) program. K.L.W. and J.A.R. would like to acknowledge earlier support for wastewater monitoring from the University of California Office of the President (award R00RG2814) and the Hewitt Foundation for Biomedical Research. We thank the City of Los Angeles and the Hyperion Wastewater Reclamation Plant for sampling assistance. We also thank Seung-Ah Chung and Melanie Oaks of the University of California Irvine Genomics Research and Technology Hub (GRT Hub), parts of which are supported by NIH grants to the Comprehensive Cancer Center (P30CA-062203) and the UCI Skin Biology Resource Based Center (P30AR075047) at the University of California, Irvine, as well as to the GRT Hub for instrumentation (1S10OD010794-01 and 1S10OD021718-01).
Author information
Authors and Affiliations
Contributions
J.T.K. and J.A.R. conceived the study; K.L., J.A.S. and J.F.G. collected wastewater samples; J.A.R. ran sequencing experiments; J.T.K. imported sequencing data, with processing and analysis by S.L.G. and W.J.B. S.L.G. wrote the manuscript, with feedback from all authors. K.L.W. provided study design, project management, and oversight along with manuscript edits.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Grimm, S.L., Rothman, J.A., Bradshaw, W.J. et al. Deep metatranscriptomic sequencing data of wastewater from Los Angeles, USA, 2023–2024. Sci Data (2025). https://doi.org/10.1038/s41597-025-06475-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-06475-7