Abstract
AML is a genetically heterogeneous disease and understanding how different co-occurring mutations cooperate to drive leukemogenesis will be crucial for improving diagnostic and therapeutic options for patients. MIR142 mutations have been recurrently detected in IDH-mutated AML samples. Here, we have used a mouse model to investigate the interaction between these two mutations and demonstrate a striking synergy between Mir142 loss-of-function and IDH2R140Q, with only recipients of double mutant cells succumbing to leukemia. Transcriptomic analysis of the non-leukemic single and leukemic double mutant progenitors, isolated from these mice, suggested a novel mechanism of cooperation whereby Mir142 loss-of-function counteracts aberrant silencing of Hoxa cluster genes by IDH2R140Q. Our analysis suggests that IDH2R140Q is an incoherent oncogene, with both positive and negative impacts on leukemogenesis, which requires the action of cooperating mutations to alleviate repression of Hoxa genes in order to advance to leukemia. This model, therefore, provides a compelling rationale for understanding how different mutations cooperate to drive leukemogenesis and the context-dependent effects of oncogenic mutations.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) arises from the accumulation of mutations in hematopoietic stem or progenitor cells resulting in the formation of an aberrant leukemic clone with defective differentiation. The expansion of this clone leads to impaired hematopoiesis and, subsequently, bone marrow failure. AML is a heterogeneous disease and a large number of recurrently mutated genes have been identified, with most leukemia cells carrying at least two driver mutations, but it is not known how these mutations collaborate to drive leukemogenesis1.
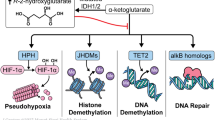
One set of frequently mutated genes are the isocitrate dehydrogenase (IDH) genes. Mutations in either IDH1 or IDH2 are common in AML, found in about 20% of cytogenetically normal patients, and occur at three particular residues—R132 in IDH1, and R140 or R172 in IDH2. The mutations at these residues disrupt the ability of the IDH enzymes to convert isocitrate to α-ketoglutarate2, and, importantly, display neomorphic activity, resulting in the aberrant production of high levels of the oncometabolite R-2-hydroxyglutarate (2-HG), which inhibits the function of dioxygenases. α-ketoglutarate-dependent dioxygenases are a broad group which includes epigenetic modifiers such as the TET family of enzymes, involved in DNA demethylation, and Jumonji-domain-containing histone demethylases3,4,5,6.
In the Cancer Genome Atlas (TGCA) AML cohort, four patients out of 200 were identified with mutations in the miRNA MIR142, all of which were exclusively identified in IDH-mutated AML samples, suggesting a strong genetic cooperation between these two types of mutation1. Three patients had an IDH2R140 mutation while one had an IDH1R132 mutation. In addition, MIR142 was found to be recurrently mutated in another cohort of AML and MDS patients, as well as in some lymphomas7,8.
MIR142 is a highly conserved miRNA which is abundantly expressed, predominantly in the hematopoietic system, and which regulates the differentiation and function of multiple hematopoietic cell types including megakaryocytes9, CD4+ dendritic cells10, T cells11,12,13,14, and erythrocytes15. The mutations in MIR142 found in AML and MDS were exclusively located in the miR-142-3p region (Fig. 1a), consistent with the observation that this is the predominant mature form of MIR142 in hematopoietic cells10,16. Furthermore, all variants affected the seed sequence of miR-142-3p and are thus likely to disrupt the interaction between miR-142-3p and its cognate targets.
Mutations affecting the seed region of miR-142-3p identified in AML patients result in loss of function. (a) Mir142 pre-miR hairpin showing position and nature of individual single nucleotide variants identified in AML and MDS patients. Numbering based on position in pre-miR sequence. Mature miRNA sequences shown in bold with seeds underlined. Wild type miR-142-3p is in green with mutations identified by TCGA in red and those identified by Thol et al. in blue. (b) Luciferase assay showing relative expression of a miR-142-3p reporter construct in cells transduced with empty vector (negative control), WT MIR142 vector (positive control), or vectors expressing MIR142 variants corresponding to each of the three TCGA point mutations—mut55 A > G, mut57 U > C and mut58 G > C. Numbering based on position in pre–miR sequence. Data shown as mean ± standard deviation of relative luminescence activity of firefly luciferase normalized to Renilla luciferase (n = 3).
The striking co-incidence of mutant miR-142-3p with neomorphic IDH1/2 mutations strongly suggested that MIR142 and IDH mutations are likely to exhibit a synergistic leukemogenic effect. A recent study investigated the potential interaction between these mutations, but synergy between IDH2R172 and loss of Mir142 was obscured by the ability of IDH2R172 alone to drive the development of a fatal myeloid neoplasia17. However, the MIR142 mutations identified in the TCGA cohort co-occurred with either IDH2R140 or IDH1R132, but not IDH2R172 (ref.1). IDH2R172 alleles have been suggested to be stronger than IDH2R140, with increased production of 2-HG18,19, which is consistent with the observation that mutations at this site do not significantly co-occur with other recurrent mutations, and IDH2R172 mutant-leukemias have been classified into a distinct genetic sub-group20,21,22,23. In contrast, IDH2R140 and IDH1R132 mutants often co-occur with other recurrent mutations, particularly NPM121,22. Mouse models have revealed that IDH2R140 and IDH1R132 variants are insufficient to drive leukemogenesis on their own but are able to do so when introduced in combination with other oncogenes commonly associated with AML24,25,26. However, many of the mutations investigated in these studies are not reflective of those that naturally co-occur with mutant IDH in human leukemias and the mechanism underlying the requirement for cooperating mutations in these IDH-dependent leukemias has yet to be elucidated.
Here, we have investigated the mechanism of cooperation between mutations in two genes that are co-mutated in AML. We combined Mir142 loss-of-function with a disease-relevant allele of IDH2, IDH2R140Q. This analysis revealed that Mir142 loss-of-function unlocked the leukemogenic potential of IDH2R140Q, and uncovered an unanticipated negative impact of IDH2R140Q upon expression of pro-leukemic Hoxa cluster genes in myeloid progenitors which was alleviated by Mir142 loss-of-function, thus releasing their combined leukemogenic potential. This study highlights the possibility that oncogenic driver mutations such as IDH2R140, can act incoherently in leukemogeneis, with progression to leukemia being dependent upon the compensatory activity of a cooperating mutation to counteract their anti-leukemic effects.
Results
Mutations in miR-142-3p identified in AML cause loss of function
MIR142 is the only miRNA found to be recurrently mutated in AML and, significantly, all the mutations so far identified were point mutations localized at multiple different positions within the seed region of miR-142-3p (Fig. 1a). Since the seed region of miRNAs is the major determinant for binding to cognate targets, it is likely that these mutations cause loss of function. To investigate this, we examined the effect of the variant forms of MIR142 on the expression of a luciferase reporter containing three copies of a bulged miR-142-3p canonical binding site. HEK293T cells were transduced with lentiviral vectors expressing either WT MIR142 or MIR142 variant hairpins corresponding to each of the three TCGA point mutations—mut55 A > G, mut57 U > C and mut58 G > C (numbering based on position in the pre-miR hairpin as shown in Fig. 1a), or empty vector control. The activity of the luciferase reporter was reduced by WT MIR142 expression, but the variant forms all failed to cause downregulation (Fig. 1b). These findings were consistent with those reported by others8,17 and demonstrated that the MIR142 mutations identified in leukemia patients result in loss of function, providing a strong rationale for using Mir142 knockout (KO) mice to investigate the role of MIR142 mutations in AML etiology.
Mir142 loss-of-function synergises with IDH2R140Q to promote leukemogenesis in a mouse model
To investigate whether Mir142 loss-of-function and IDH2R140Q have a synergistic leukemogenic effect, we used two strains of Mir142 KO mice (Supplementary Fig. 1a,b). CRISPR-Cas9 genome editing was used to generate a specific 26 bp deletion of the region encoding miR-142-3p (Mir142em2Card, provided by Masatake and Kimi Araki, Centre for Animal Resources and Development (CARD), Kumamoto University, Japan), and we confirmed our findings using the Mir142HOJ2 strain which harbors a 276 bp deletion of the Mir142 locus generated through germline Cre-mediated recombination of a LoxP targeted allele (provided by Graham Lord, King’s College London, UK, Supplemental Fig. 1a)14. Loss of Mir142 expression was confirmed by qRT-PCR (Supplementary Fig. 1b). Both the Mir142HOJ2 and Mir142em2Card strains exhibited the same phenotypes, and we refer to them collectively as Mir142−/− mice. Consistent with the findings of Trissal et al., Mir142−/− mice did not develop leukemia, demonstrating that Mir142 loss-of-function alone is not sufficient for leukemogenesis17.
To investigate whether IDH2R140 mutants cooperate with Mir142 loss-of-function to promote leukemic transformation, we introduced the IDH2R140Q gene into Mir142−/− and wild-type hematopoietic stem/progenitor cells (HSPCs) using a lentiviral construct co-expressing the oncogene together with a GFP reporter (Fig. 2a, Supplementary Fig. 1c,d). FACS sorted CD45.2+ KLS HSPCs were transduced with SFFV-IDH2R140Q-IRES-GFP (IDH2R140Q) or SFFV-IRES-GFP control (CTL) vector and transplanted into irradiated CD45.1+ recipients with WT CD45.1 + whole bone marrow (BM) competitor cells (Fig. 2a). Consistent with the reported neomorphic function of IDH mutants, we observed high levels of 2-HG in IDH2R140Q-expressing BM cells (Supplementary Fig. 1e). Strikingly, all the recipients of Mir142−/− cells transduced with IDH2R140Q vector, (Mir142−/− + IDH2R140Q double mutants) became ill, suffering from severe cytopenia and anemia, and had to be culled (median survival = 8 months; Fig. 2b–e). In contrast, all control animals transplanted with WT cells transduced with CTL vector (WT + CTL), and the majority of recipients transplanted with WT cells transduced with IDH2R140Q (WT + IDH2R140Q), remained healthy throughout the experiment. There was, therefore, a clear synergistic impact on survival resulting from the combination of Mir142 loss-of-function with IDH2R140Q compared to IDH2R140Q alone.
Mir142 loss of function synergizes with IDH2R140Q to promote myeloid leukemogenesis in mice. (a) Schematic diagram summarizing the model. KLS: c-Kit+ Lin- Sca1+ cells. (b) Kaplan–Meier analysis of survival of mice transplanted with WT + CTL (empty vector control, black, n = 5), WT + IDH2R140Q (red, n = 7) or Mir142 –/– + IDH2R140Q (blue, n = 8) HSPCs. Mantel-Cox log–rank test: Mir142 –/– + IDH2R140Q versus WT CTL P = 0.0308, Mir142 –/– + IDH2R140Q versus WT + IDH2R140Q P = 0.0134, and WT + IDH2R140Q versus WT CTL P = 0.617 (not significant). Survival data from 2 independent experiments using the two different Mir142–/– strains. (c,d) Red blood cell (RBC), (c) and white blood cell counts (WBC), (d) from recipients of transduced HSPCs, analyzed 4–6 months after transplantation (WT + CTL n = 8, WT + IDH2R140Q n = 11, Mir142 –/– + IDH2R140Q n = 13). (e) Hemoglobin levels in peripheral blood of transplant recipients 4–6 months after transplantation (WT + CTL n = 7 , WT + IDH2R140Q n = 9, Mir142 –/– + IDH2R140Q n = 10). Data shown in c-e pooled from 3–4 independent experiments, using both Mir142–/– strains (Mir142em2Card: open blue circles. Mir142HOJ2: filled blue circles).
Impaired hematopoiesis and expansion of myeloid progenitors in Mir142 KO mice
To understand the relative contributions of Mir142 and IDH2 mutations to leukemogenesis, we examined the hematopoietic defects associated with loss of Mir142 alone using our KO mouse models. Analysis of peripheral blood (PB) in Mir142 KO mice revealed pan-cytopenia (Fig. 3a,b) arising from a reduction in all lineages (Fig. 3c). Transplantation of wild type or Mir142−/− BM or HSPCs (CD45.2+) into irradiated CD45.1+ recipients revealed that Mir142−/− HSCs exhibit reduced reconstitution capacity (Fig. 3d) suggesting that the defect is intrinsic to the hematopoietic compartment and not due to impairment of the BM niche in KO mice. Mir142−/− mice also exhibited significant splenomegaly, with a doubling in spleen mass (Fig. 3e), and there was a large increase in the Mir142−/− Mac1+Gr1+ myeloid population in the spleens of both the constitutive Mir142 KO and the transplant recipients (Fig. 3f,g). Splenomegaly and extramedullary hematopoiesis can be associated with inefficient myelopoiesis and so may reflect a compensatory response to the reduction in circulating myeloid cells observed in the Mir142−/− mice.
Mir142 loss of function leads to impaired hematopoiesis and reduced hematopoietic reconstitution capacity. (a,b) WBC (a) and RBC (b) counts from WT and Mir142–/– mice (WT n = 8, Mir142–/– n = 9). (c) Lineage distribution within peripheral blood WBCs (WT n = 4, Mir142–/– n = 5). (d) Proportion of CD45.2+ donor cells within total CD45 (CD45.1 + CD45.2) in peripheral blood at 3 months after transplantation of WT or Mir142–/– HSPCs (WT n = 4, Mir142–/– n = 3). (e) Spleen size in WT and Mir142–/– mice (n = 11 each). (f) Lineage distribution within CD45+ splenocytes from WT and Mir142–/– mice: B cells (B220+), T cells (CD3+), granulocytes (Mac1+Gr1+), megakaryocytes (CD41+) and erythroid progenitors (CD71+Ter119+) (n = 5 each for WT and Mir142–/–). (g) Frequency of Mac1+Gr1+ myeloid cells within CD45.2+ donor cells in the spleens of recipients transplanted with WT or Mir142–/– bone marrow cells (WT n = 5, Mir142–/–n = 4). Pooled data is shown in (a,b), and (e), from analysis of both strains of Mir142–/– mice (Mir142HOJ2 : open blue circles. Mir142em2Card: filled blue circles). Data in (c,g) generated with Mir142HOJ2, and data in (d,f) generated with Mir142em2Card.
The proportion of HSCs, defined as either CD34−Flt3-KLS (LT-HSC) or CD150+CD48-KLS (SLAM HSC), was not significantly impacted in the Mir142−/− mice (Fig. 4a); however, there was a significant expansion of the KLS population, predominantly accounted for by increased numbers of multipotent progenitors defined as either CD34+Flt3-KLS (ST-HSC) or CD150-CD48+ (SLAM HPC1) progenitors (Fig. 4a). Analysis of the committed progenitor compartment revealed myeloid skewing in Mir142−/− mice, with a significant increase in the proportion of GMPs, and a concomitant reduction of MEPs in the bone marrow of the KO mice (Fig. 4b). There was also a large increase in the number of GMPs in the spleens of the KO mice and within the donor-derived Mir142−/− cells in the spleens of the transplant recipients (Fig. 4c,d). In addition, there was a significant increase in the proportion of myeloid colonies generated by Mir142−/− HSPCs in CFC assays, consistent with the myeloid-bias observed in the progenitor populations in these animals (Fig. 4e).
Mir142 loss of function leads to expansion of myeloid progenitors and formation of an aberrantly expanded CD34+Mac1+ myeloblast population. (a) Proportion of HSC subsets within the bone marrow as assessed by CD34/Flt3 or SLAM markers (CD150/CD48) (WT n = 10, Mir142–/– n = 9). LT-HSC (CD34–Flt3– KLS), ST-HSC (CD34+ Flt3– KLS), LMPP (CD34+ Flt3+ KLS), SLAM HSC (CD150+ CD48– KLS), SLAM MPP (CD150– CD48– KLS) and SLAM HPC1 (CD150–CD48+ KLS). (b) Proportion of myeloid progenitors within the bone marrow (WT n = 10, Mir142–/– n = 9). (c) Proportion of multipotent and committed progenitor populations in the spleen (WT n = 9, Mir142–/– n = 8). (d) Frequency of multipotent (KLS) and myeloid committed progenitors (GMPs) within CD45.2+ donor cells in the spleens of transplant recipients (WT n = 5, Mir142–/– n = 4). (e) Proportion of myeloid, megakaryocyte/erythroid (Meg/E) and mixed colonies in CFC assay from FACS sorted HSPCs (KLS). Pooled data shown from 4 independent experiments. Statistical significance shown for proportion of myeloid colonies. (f,g) Proportion of CD34+ Mac1+ cells within CD45+ bone marrow cells (f) and CD45+ splenocytes (g) (n = 5 each for WT and Mir142–/–). (h) Proportion of CD34+Mac1+ cells within CD45.2+ donor cells in the bone marrow of recipients transplanted with WT or Mir142–/– HSPCs (WT n = 4, Mir142–/– n = 3). All data generated using Mir142em2Card KO mice, except D, for which the Mir142HOJ2 strain was used.
However, the clearest abnormality was the presence of an aberrant myeloblast population in the bone marrow and spleens of the Mir142−/− animals and in recipients transplanted with Mir142−/− cells. This population co-expressed Mac1+, a marker of committed myeloid cells, and CD34, a protein normally confined to primitive hematopoietic stem and progenitor cells, suggesting a partial block in myeloid differentiation (Fig. 4f–h) consistent with the reduction in circulating Mac1+Gr1+ cells in the Mir142−/− mice. Increased frequency of myeloblasts is a defining feature of AML, and the presence of this expanded CD34+ Mac1+ aberrant myeloid population in Mir142−/− animals strongly supported a role for Mir142 loss-of-function in promoting leukemogenesis; however, this population never rose above 10% of CD45+ cells in the bone marrow, consistent with the failure of Mir142 loss-of-function to promote leukemia in the absence of IDH2R140Q.
Cooperation between Mir142 loss-of-function and IDH2R140Q drives leukemic transformation of myeloid progenitors
To investigate how Mir142 and IDHR140 mutations synergize to drive leukemogenesis, we further explored the effect of IDH2R140Q in the presence or absence of Mir142. There was a significant increase in the proportion of GFP+ donor cells in the bone marrow of Mir142−/− + IDH2R140Q recipients compared to the recipients of wild type cells transduced with empty vector (WT + CTL), such that the majority of cells in the bone marrow of the double-mutant recipients were GFP+, demonstrating that the combination of Mir142 loss-of-function and IDH2R140Q provides a strong competitive advantage (Fig. 5a). In contrast, there was no significant increase in the frequency of GFP+ cells in the recipients of single mutant WT + IDH2R140Q cells. Notably, Mir142−/− + CTL HSPCs failed to engraft (< 1% donor chimerism) precluding further analysis, but transduction of Mir142−/− cells with IDH2R140Q rescued their defective engraftment and allowed their expansion in the recipients, resulting in the development of leukemia (Fig. 5a). Therefore, the IDH2R140Q mutant compensated for a detrimental effect of Mir142 loss-of-function in this setting.
Expansion of myeloid progenitors and CD34+Mac1+ myeloblasts in Mir142–/– + IDH2R140Q leukemias. (a) Proportion of GFP+ cells in the bone marrow from recipients of transduced HSPCs (WT + CTL n = 7, Mir142–/– + CTL n = 5, WT + IDH2R140Q n = 9, Mir142–/– + IDH2R140Q n = 10). (b) Proportion of Mac1+ cells within CD45.2+GFP+ population in peripheral blood from recipients of transduced HSPCs (WT + CTL n = 9, WT + IDH2R140Q n = 12, Mir142 –/– + IDH2R140Q n = 14). Arrow indicates an animal which had a lower proportion of Mac1 + cells due to the majority (66%) of CD45.2+GFP+ peripheral blood cells having a more primitive CD34+Mac1– phenotype. (c) Percentage GFP+ cells in spleens from recipients of transduced HSPCs (WT + CTL n = 5, WT + IDH2R140Q n = 7, Mir142 –/– + IDH2R140Q n = 9). (d) Proportion of GFP+ cells in the livers from recipients of transduced HSPCs (WT + IDH2R140Q n = 4, Mir142 –/– + IDH2R140Q n = 4). (e) Spleen size in recipients of transduced HSPCs (WT + CTL n = 7, WT + IDH2R140Q n = 11, Mir142 –/– + IDH2R140Q n = 13). (f,g) Proportion of GMPs within GFP+ progenitors (c-Kit+ Lin– Sca1– compartment) in the bone marrow (f) or spleens (g) of recipients of transduced HSPCs (WT + IDH2R140Q n = 6, Mir142–/– + IDH2R140Q n = 8). (h) Representative FACS plots showing proportion of CD34+ Mac1+ myeloblasts within the GFP+ population in the bone marrow (BM, top panels) or spleen (SP, lower panels) from recipients of transduced HSPCs. (i,j) Proportion of CD34+ Mac1+ myeloblasts within the GFP+ population in the bone marrow (i) or spleen (j) from recipients of transduced HSPCs (WT + IDH2R140Q n = 6, Mir142–/– + IDH2R140Q n = 8). (k) Cytospin of Mir142–/– + IDH2R140Q CD34+Mac1+ cells showing myeloblastic morphology. Data in (a–c,e) is pooled from 3 to 4 independent experiments using both Mir142em2Card (open circles) and Mir142HOJ2 (filled circles) KO mice. Data in (f–j) generated using only the Mir142em2Card allele, pooled from 2 independent experiments.
In line with the synergistic impacts seen on survival and on inhibition of normal hematopoiesis (severe cytopenia and anemia) (Fig. 2b–e), the Mir142−/− + IDH2R140Q recipients displayed an increased proportion of Mac1+ myeloid cells in the peripheral blood, consistent with myeloid neoplasia (Fig. 5b). Furthermore, GFP+ leukemia cells infiltrated into the spleen and liver (Fig. 5c,d), and double mutant (Mir142−/− + IDH2R140Q) recipients exhibited more severe splenomegaly compared to both the single mutant (WT + IDH2R140Q) recipients (Fig. 5e) and the Mir142 knockout alone (Fig. 3e).
AML is a heterogeneous disease characterized by a block in myeloid differentiation and expansion of aberrant myeloid progenitors. Many studies have demonstrated that AML may arise from transformation of committed myeloid progenitors27,28,29,30. In the leukemias generated by Mir142−/− + IDH2R140Q cells, the majority of the lineage negative, c-Kit+ progenitor cells in the bone marrow had a GMP-like phenotype, and the proportion of GMPs was significantly increased in the Mir142−/− + IDH2R140Q double mutants compared to the single IDH2R140Q mutants (48% compared to 21% of GFP+ BM progenitor cells) (Fig. 5f), with a particularly large increase in the frequency of GMPs within the spleen (Fig. 5g). In addition, there was a very large (~ sixfold) expansion in the number of CD34+Mac1+ myeloblasts in the leukemic Mir142−/− + IDH2R140Q recipients (Fig. 5h–j) compared to single mutant WT + IDH2R140Q recipients (from 4% to 25% of GFP+CD45.2+ BM cells). This was also significantly higher than observed for Mir142 loss-of-function alone, in either steady state or transplantation settings (Fig. 4f–h). Cytospins of FACS-purified CD34+Mac1+ cells confirmed that this population was composed of primitive myeloid cells with a myeloblastic morphology (Fig. 5k). This analysis therefore revealed a strong synergistic impact of Mir142 loss-of-function and IDH2R140Q in promoting expansion of immature myeloid progenitor populations, underlying their cooperative leukemogenic effect.
Mir142 loss-of-function antagonizes IDH2R140Q-dependent silencing of Hoxa cluster genes in myeloid progenitors
To investigate the mechanism of cooperation between Mir142 loss-of-function and IDH2R140Q, we performed RNA-seq analysis on myeloid progenitors (GMPs) isolated from WT and Mir142−/− mice, as well as WT + CTL, WT + IDH2R140Q, and Mir142−/− + IDH2R140Q GMPs isolated from transplant recipients (Fig. 6a, Supplementary Fig. 2a–c, Supplementary data files 1–3).
Mir142 loss of function upregulates a leukemic HOX/Meis1/Pbx3 signature and antagonizes IDH2R140Q-dependent silencing of Hoxa cluster genes. (a) Volcano plot showing differentially expressed genes (DEGs) in Mir142–/– versus WT GMPs (padj < 0.05 colored dots, WT n = 4, Mir142–/– n = 3). miR-142-3p targets predicted by Targetscan highlighted in blue. Light blue: miR–142–3p.1 specific targets. Dark blue: miR-142-3p.2 specific targets. Turquoise: targets of both miR-142-3p.1 and miR-142-3p.2 isomirs. Top ten most significantly differentially expressed genes are labelled. (b) Gene set enrichment analysis (GSEA) reveals strong enrichment of predicted miR-142-3p targets (union of miR-142-3p.1 and miR-142-3p.2 targets) in genes upregulated in Mir142–/– GMPs compared to WT. (c) Venn diagram showing overlap of miR-142-3p.1 and miR-142-3p.2 targets with DEGs from Mir142–/– versus WT GMPs and Mir142–/– + IDH2R140Q versus WT + IDH2R140Q GMPs (all DEGs with padj < 0.05 were used for the analysis). Highlighted targets Ash1l and Ctnnb1 are differentially expressed in both Mir142–/– and Mir142–/– + IDH2R140Q GMPs. (d) GSEA reveals strong enrichment of a mutant NPM1–associated leukemic signature in genes upregulated in Mir142–/– + IDH2R140Q GMPs compared to WT + IDH2R140Q. (e) Heatmap showing expression of the significantly differential leading-edge genes contributing to enrichment of the mutant NPM1–associated leukemic signature in Mir142–/– + IDH2R140Q verus WT + IDH2R140Q GMPs (padj < 0.05). Signature includes multiple homeobox family genes (Hoxa5/7/9/10, Meis1 and Pbx3) highlighted with arrows. Expression in Mir142–/–GMPs is shown for comparison. Columns represent normalized expression values from each sample isolated from individual mice (WT + IDH2R140Q (n = 4), Mir142–/– + IDH2R140Q (n = 6), and Mir142–/– (n = 3)). (f) Heatmap of most significantly differentially expressed genes in CD34+Mac1+ myeloblasts and Mac1+(CD34–) myeloid cells isolated from Mir142–/– mice and Mir142–/– + IDH2R140Q recipients (GFP + cells). (g) Expression of differentially expressed Hoxa cluster genes and Meis1 in CD34+Mac1+ myeloblasts and Mac1+(CD34–) myeloid cells in bone marrow of Mir142–/– mice (padj < 0.05, n = 2). (h) Expression of differentially expressed Hoxa cluster genes and Meis1 in Mir142–/– + IDH2R140Q GMPs, CD34+Mac1+ myeloblasts and mature Mac1+(CD34–) cells (padj < 0.05, n = 2). (i) Expression of homeobox genes including Hoxa cluster genes (Hoxa5/7/9/10), Meis1 and Pbx3, and the HOX regulator Ash1l in WT + CTL (n = 3), WT + IDH2R140Q (n = 4), Mir142–/– + IDH2R140Q (n = 6) and Mir142–/– (n =3) GMPs. (j) CFC assay with WT and Mir142–/– HSPCs co-transduced with IDH2R140Q and either an shRNA targeting Ash1l (shAsh1l) or non-targeting control (shNeg) (n = 2). Statistical significance assessed by unpaired two-tailed t test.
Predicted targets of miR-142-3p were highly enriched within the genes upregulated in Mir142−/− and double mutant Mir142−/− + IDH2R140Q cells, accounting for 16% and 17% of upregulated genes respectively (Fig. 6a–c, Supplementary data file 1). miR-142-3p has been reported to produce two isomirs (miR-142-3p.1 and miR-142-3p.2) which differ by one nucleotide at the 5′ end31,32. Predicted targets of both isomirs were highly enriched within the upregulated genes in both Mir142−/− and Mir142−/− + IDH2R140Q cells compared to WT or WT + IDH2R140Q controls respectively, (Fig. 6a–c, Supplementary Fig. 2b,h) indicating that both isomirs are functional in myeloid progenitors.
Gene set enrichment analysis revealed a strong enrichment for a mutant-NPM1 leukemia associated signature within the genes upregulated in the double mutant (Mir142−/− + IDH2R140Q) GMPs, including several homeobox genes and HOX cofactors33 (Fig. 6d,e). The Hoxa cluster genes—Hoxa5, Hoxa7, Hoxa9, and Hoxa10—and genes encoding the leukemogenic Hox co-factors, Meis1 and Pbx3, were all upregulated in double mutant Mir142−/− + IDH2R140Q GMPs compared to their WT + IDH2R140Q counterparts (Fig. 6e, Supplementary data file 2). In addition, Ash1l and Ctnnb1 (β-catenin), which have previously been shown to mediate expansion of myeloid progenitors and promote leukemic transformation in MLL/HOX-driven leukemias34,35,36, were both significantly upregulated in Mir142−/− + IDH2R140Q GMPs (Fig. 6c,i, Supplementary Fig. 2d and Supplementary data file 2). Ash1l, Ctnnb1, Hoxa7, and Hoxa9 were also upregulated in the single mutant Mir-142−/− GMPs compared to WT, but the key co-factors Meis1 and Pbx3 were not, potentially explaining why these mice failed to develop leukemia (Supplementary data file 1).
To understand the origin of the aberrantly expanded CD34+ Mac1+ myeloblast population we isolated this population, together with the CD34- Mac1+ cells from both the leukemic Mir142−/− + IDH2R140Q recipients and the single mutant Mir142−/− mice, and performed RNA-seq (Fig. 6f). This analysis revealed that the Mir142−/− CD34+ Mac1+ myeloblasts expressed very high levels of Hoxa cluster genes compared to the more differentiated CD34- Mac1+ myeloid cells (Fig. 6g). Furthermore, in leukemic Mir142−/− + IDH2R140Q recipients, both the CD34+ Mac1+ and CD34- Mac1+ myeloid cells expressed high levels of Hoxa genes, similar to the levels of expression in GMPs (Fig. 6h). The expansion of myeloid populations in Mir142−/− animals and in Mir142−/− + IDH2R140Q leukemias is therefore likely to be caused by failure to downregulate Hoxa cluster genes during myeloid differentiation.
Given our observations, and the oncogenic nature of IDH2 mutations, we initially expected that Hoxa genes would be activated by IDH2R140Q; however, to our surprise, we discovered that Hoxa5, Hoxa7 and Hoxa9 were in fact expressed at a significantly lower level in IDH2R140Q GMPs (Fig. 6i and Supplementary data file 3). This effect was specifically observed in myeloid progenitors and not in KLS HSPCs (Supplementary Fig. 2e). Crucially, this suppression of Hoxa cluster expression was alleviated in Mir142−/− + IDH2R140Q GMPs (Fig. 6i), and Hoxa10 was significantly upregulated, revealing that loss of Mir142 function cooperates with IDH2R140Q by preventing downregulation of these key regulators of leukemic transformation. This led us to speculate that it may be a pre-requisite for mutations that collaborate with IDH2R140 mutants to counteract Hoxa cluster repression.
Hoxa cluster activation through a Mir142-Ash1l axis is required for expansion of myeloid progenitors
We then asked how Mir142 loss-of-function promotes Hoxa cluster activation, and whether upregulation of Hoxa genes is required for the expansion of myeloid progenitors in the Mir142−/− + IDH2R140Q cells. We first identified predicted miR-142-3p targets that were differentially expressed both in Mir142−/− and in Mir142−/− + IDH2R140Q GMPs (Fig. 6c). Of these predicted targets, an obvious candidate was Ash1l, which encodes a histone H3K36 methyltransferase belonging to the MLL/Trithorax family, which has important functions in promoting HOX gene expression in both normal and leukemic cells36,37,38,39. Targetscan predicted four miR-142-3p target sites in the Ash1l 3′UTR—two of which are highly conserved40. Ash1l has been previously shown to be a direct target of miR-142-3p, and depletion of Ashl1 using a gene trap allele prevented upregulation of Hoxa9 in Mir142−/− bone marrow17,41.
We therefore investigated whether the knockdown of Ash1l could ameliorate the effect of Mir142 loss-of-function in myeloid progenitors using an shRNA targeting Ash1l (Fig. 6j, Supplementary Fig. 2f,g). Mir142−/− + IDH2R140Q HSPCs produced an increased proportion of myeloid colonies compared to WT + IDH2R140Q HSPCs (Fig. 6j). However, knockdown of Ash1l partially suppressed this increase in myeloid colony formation, while having no significant effect on WT + IDH2R140Q cells or Mir142−/− + CTL cells (Fig. 6j and Supplementary Fig. 2g). This data is consistent with a compensatory antagonistic model of cooperation between these mutations, wherein Mir142 loss-of-function counteracts the suppression of HOX gene expression by IDH2R140Q, through the upregulation of HOX regulatory factors such as Ash1l, thus unlocking leukemic transformation of IDH2R140Q-mutated myeloid progenitors.
Discussion
AML is a highly intractable cancer, and the prognosis for AML patients has improved little in recent years despite significant increases in our understanding of the genetic and epigenetic complexity of this disease. The key to developing better treatments will be to understand how the different recurrent mutations identified in AML patients collaborate to drive development of overt leukemia. However, this will require careful modelling of each these co-occurring mutations, individually and in combination, to identify the underlying mechanisms of their cooperative leukemogenic effects.
Here, we have modelled the synergy between two disease-relevant, co-occurring mutations in an in vivo setting, and used gene expression analysis of defined progenitor subsets to investigate how they cooperate to drive leukemogenesis. The MIR142 mutations identified in AML patients, exclusively co-occurred with IDH mutations in the TCGA cohort, and in one patient from the cohort of AML and MDS patients described by Thol et al.1,8. Our analysis revealed that, counterintuitive to its oncogenic role, IDH2R140Q actually leads to lower expression of the pro-leukemic Hoxa cluster genes in GMPs but this is counteracted by the loss of Mir142. Recipients of Mir142−/− + IDH2R140Q double mutant HSPCs developed a myeloid leukemia characterized by an expansion of immature myeloid cells and a profound inhibition of normal hematopoiesis resulting in peripheral cytopenia, that was not induced by either Mir142 loss-of-function or IDH2R140Q alone; thus demonstrating that loss of Mir142 unlocks the leukemogenic potential of IDH2R140Q. Knockdown experiments suggested that this effect is likely to be mediated, at least in part, by the upregulation of the MLL/Trithorax-family histone methyltransferase ASH1L, a known regulator of HOX gene expression in both normal and leukemic cells.
The mutations affecting MIR142 in AML were all located in the seed region of miR-142-3p suggesting that they cause loss of targeting of canonical MIR142 targets and we and others have confirmed this using luciferase reporter assays8,17. However, it is possible that the mutated seed can recognise new targets and could contribute to the impact of these mutations. However, since the mutations are located at 5 different positions, each would create a different neo-target making it unlikely that they have convergent effects. It is therefore likely that loss of function is the main cause of the leukemogenic function of these mutants as supported by the finding by us and others that Mir142 loss synergises with IDH2 mutations to promote leukemogenesis in mice17.
Activation of the HOX cluster is a key feature of many types of AML, and HOXA9 is a potent oncogene42,43,44,45,46. Nevertheless, HOXA9 requires the upregulation of the homeodomain-containing co-factors, MEIS1 or PBX3, to promote leukemogenesis47,48,49,50. While Hoxa genes were upregulated in Mir142−/− GMPs, Meis1 and Pbx3 were not, consistent with the failure of these mice to develop leukemia. However, Meis1 and Pbx3 were activated in Mir142−/− + IDH2R140Q leukemic GMPs, suggesting that synergistic activation of these co-factors may contribute to the leukemogenic effect of our combined mutations in Mir142 and IDH2. Our data, therefore, points to a dual mechanism underpinning the inter-dependence of Mir142 loss-of-function and IDH mutations in AML: antagonistic regulation of HOXA cluster expression and mutual activation of homeobox co-factors.
The antagonistic effect of Mir142 and IDH on HOX gene expression may have wider implications outside of this small subgroup of Mir142 mutant AML, as it suggests that IDH mutations—which are one of the most common aberrations in AML—may stringently require HOX activation for their leukemogenic consequences to be realized. Notably, IDH mutations frequently co-occur with NPM1 mutations, which are strongly associated with a HOX gene expression signature33,51, and HOX/Meis1 overexpression is required to maintain the leukemic state in NPM1 mutant cells52. Therefore mutations in NPM1 and Mir142 may provide a convergent role in IDH-mutant leukemias, namely to activate HOX gene expression.
In summary, our findings provide a new framework for understanding genetic cooperativity in cancer. The prevailing concept is that each and every consequence of a cancer-associated mutation directly contributes to increased risk of tumour formation or, at least, is neutral in its current context. However, multistep tumorigenesis and clonal evolution may be better understood in terms of less coherent outcomes from mutations. We propose that cancer-associated variants can act incoherently, with both positive and negative effects on oncogenesis, resulting in the co-selection of mutations that alleviate their tumour-suppressive properties. It will be interesting to identify further examples of this type of genetic complementation between co-occurring driver mutations. This model provides a compelling rationale for understanding the context-dependency of so-called driver mutations which are often necessary, but not sufficient, to drive oncogenesis.
Methods
Animals
Mice were maintained in specific pathogen-free conditions and all experimental protocols were performed in accordance with United Kingdom Home Office regulations. Two mouse models with Mir142 deletions were used in this study. The B6-Mir142em2Card strain carries a 26 bp deletion of the miR-142-3p sequence generated by CRISPR-Cas9 editing (provided by Masatake and Kimi Araki, Centre for Animal Resources and Development (CARD), Kumamoto University, Japan). The Mir142HOJ2 line carries a 276 bp deletion of the Mir142 locus generated through germline Cre-mediated recombination of a floxed Mir142 allele (provided by Graham Lord, King’s College London, UK)14. In the transplantation studies, 8–16 week old, male and female B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice were used as recipients and to provide support/competitor cells to ameliorate effects of myeloablation. Maintenance of the B6-Mir142em2Card and CD45.1 mice, and all transplantation experiments were performed at University College London (UCL) under UK Home Office Project License PPL:70/8143. The Mir142HOJ2 line was maintained at Kings College London under PPL:70/7869.
Lentiviral vector production
VSV-G pseudotyped lentiviral vectors were produced by transient transfection of CSI lentiviral plasmids with psPAX2 and pMD2.G packaging plasmids into sub-confluent HEK293T cells using FuGene 6 (E2691, Promega, Madison, WI, USA). Viral supernatants were collected and concentrated by centrifugation at 50,000g for 2.5 h at 4 °C before being resuspended in IMDM. Viral titre was estimated by transducing HEK293T cells with serially diluted vector and quantifying the proportion of GFP + cells by flow cytometry.
Luciferase reporter assay for miR-142-3p activity
HEK293T cells were transduced at matched MOI with lentiviral vectors expressing either WT or mutant MIR142 with a GFP reporter, or empty vector control. GFP + cells were sorted after 3 days and transfected with the miR-142-3p luciferase reporter. Luciferase assays were then performed following the manufacturer’s protocol using Dual-Glo Luciferase Assay system (Promega, E2920) and luminescence measured on the Varioskan LUX (N16045, Thermo Fisher Scientific, Waltham, MA USA).
Transplantation assays
Whole bone marrow (5 × 105 cells) or c-Kit+ Lineage− Sca1+ (KLS) HSPCs (1 × 104 cells) from CD45.2 + donors were mixed with 1 × 105 CD45.1 + whole bone marrow support/competitor cells and injected into the tail vein of irradiated CD45.1 recipient mice. Irradiation was performed using an X-ray irradiator to deliver 800 cGy as a split dose.
For leukemia modelling, 1 × 104 KLS cells per recipient were FACS-purified from CD45.2 + wild type or Mir142−/− donors, and resuspended in StemSpan serum-free expansion medium (Stem Cell Technologies, Vancouver, BC, Canada), supplemented with 100 units/mL penicillin, 100 ng/ml streptomycin, 200 ng/mL SCF, 20 ng/mL Flt3-Ligand, and 20 ng/mL TPO. Cells were pre-incubated for 1–2 h at 37 °C in the above cytokines and transduced with lentiviral vector (either SFFV-IDH2R140Q-IRES-GFP or empty SFFV-IRES-GFP control vector) overnight at 37 °C, 5% CO2. The cells were then washed and co-injected with 2 × 105 CD45.1 whole bone marrow support/competitor cells into irradiated (800 cGy) CD45.1 recipient mice. Animals that showed no engraftment of donor cells were excluded from further analysis. Mice exhibiting declining health status were sacrificed and tissues taken for analysis.
Colony forming cell (CFC) assays
KLS cells were FACS-purified from bone marrow samples, seeded into 1.5 mL of Methocult M3434 (Stem Cell Technologies) and plated into 35 mm non-coated plates (430,588, Corning Incorporated, Corning, NY, USA). Plates were incubated for 10–14 days at 37 °C, 5% CO2. Colonies produced were counted and classified.
RNA-seq library preparation
RNA was extracted from KLS, GMP, CD34+Mac1+ and CD34-Mac1+ cells, and RNA-seq libraries were prepared using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (634,891, Takara Bio, Kusatsu, Japan), and the Nextera XT library preparation kit (FC-131-1096, Illumina, San Diego, CA, USA). Libraries were then sequenced on an Illumina NextSeq 500.
Statistical analysis
Statistical analyses including assessments of significance, variance and normality were performed using Prism 8 software (GraphPad, San Diego, CA, USA). Results are expressed as the mean ± standard error of the mean (SEM) unless otherwise indicated. Significant differences between experimental groups were determined by a two-tailed Mann–Whitney test unless otherwise indicated. Survival data was analyzed using the Mantel-Cox log-rank test. P values shown as follows: *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
Data availability
RNA-seq data generated during the current study was deposited in Array Express (https://www.ebi.ac.uk/arrayexpress/) with accession number: E-MTAB-8042.
Change history
23 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41598-021-84578-y
References
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Waitkus, M. S., Diplas, B. H. & Yan, H. Biological role and therapeutic potential of idh mutations in cancer. Cancer Cell 34, 186–195 (2018).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Chowdhury, R. et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 (2011).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Kwanhian, W. et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 1, 141–155 (2012).
Thol, F. et al. Clinical and functional implications of microRNA mutations in a cohort of 935 patients with myelodysplastic syndromes and acute myeloid leukemia. Haematologica 100, e122–e124 (2015).
Chapnik, E. et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. Elife 3, e01964 (2014).
Mildner, A. et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood 121, 1016–1027 (2013).
Kramer, N. J. et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 125, 3720–3730 (2015).
Sun, Y. et al. Mature T cell responses are controlled by microRNA-142. J. Clin. Invest. 125, 2825–2840 (2015).
Mildner, A. et al. MicroRNA-142 controls thymocyte proliferation. Eur. J. Immunol. 47, 1142–1152 (2017).
Anandagoda, N. et al. microRNA-142–mediated repression of phosphodiesterase 3B critically regulates peripheral immune tolerance. J. Clin. Invest. 129, 1257–1271 (2019).
Rivkin, N. et al. Erythrocyte survival is controlled by microRNA-142. Haematologica 102, 676–685 (2017).
Nimmo, R. et al. MiR-142–3p controls the specification of definitive hemangioblasts during ontogeny. Dev. Cell 26, 237–249 (2013).
Trissal, M. C. et al. MIR142 loss-of-function mutations derepress ASH1L to increase HOXA gene expression and promote leukemogenesis. Cancer Res. 78, 3510–3521 (2018).
Ward, P. S. et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J. Biol. Chem. 288, 3804–3815 (2013).
Janin, M. et al. Serum 2-hydroxyglutarate production in IDH1- and IDH2-mutated de novo acute myeloid leukemia: A study by the Acute Leukemia French Association group. J. Clin. Oncol. 32, 297–305 (2014).
Marcucci, G. et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J. Clin. Oncol. 28, 2348–2355 (2010).
Green, C. L. et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 118, 409–412 (2011).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Meggendorfer, M. et al. IDH1R132, IDH2R140 and IDH2R172 in AML: Different genetic landscapes correlate with outcome and may influence targeted treatment strategies. Leukemia 32, 1249–1253 (2018).
Sasaki, M. et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488, 656–659 (2012).
Chen, C. et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 27, 1974–1985 (2013).
Kats, L. M. et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 14, 329–341 (2014).
Cozzio, A. et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17, 3029–3035 (2003).
Krivtsov, A. V. et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature 442, 818–822 (2006).
Goardon, N. et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19, 138–152 (2011).
Quek, L. et al. Genetically distinct leukemic stem cells in human CD34-acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J. Exp. Med. 213, 1513–1535 (2016).
Wu, H., Ye, C., Ramirez, D. & Manjunath, N. Alternative processing of primary microRNA Transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE 4, e7566 (2009).
Manzano, M., Forte, E., Raja, A. N., Schipma, M. J. & Gottwein, E. Divergent target recognition by coexpressed 5′-isomiRs of miR-142-3p and selective viral mimicry. RNA 21, 1606–1620 (2015).
Verhaak, R. G. W. et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106, 3747–3754 (2005).
Yeung, J. et al. β-catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell 18, 606–618 (2010).
Wang, Y. et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science 327, 1650–1653 (2010).
Zhu, L. et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 6, 770–783 (2016).
Tanaka, Y. et al. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of hox gene expression. PLoS ONE 6, e28171 (2011).
Brinkmeier, M. L. et al. The histone methyltransferase gene absent, small, or homeotic discs-1 like is required for normal hox gene expression and fertility in Mice1. Biol. Reprod. 93, 121, 1–12 (2015).
Jones, M. et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J. Clin. Invest. 125, 2007–2020 (2015).
Agarwal, V., Bell, G. W., Nam, J.-W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005 (2015).
Colamaio, M. et al. miR-142–3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J. Clin. Endocrinol. Metab. 100, E59–E69 (2015).
Andreeff, M. et al. HOX expression patterns identify a common signature for favorable AML. Leukemia 22, 2041–2047 (2008).
Golub, T. R. et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286, 531–537 (1999).
Faber, J. et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 113, 2375–2385 (2009).
Ayton, P. M. & Cleary, M. L. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17, 2298–2307 (2003).
Collins, E. M. & Thompson, A. HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 62, 847–856 (2018).
Nakamura, T., Largaespada, D. A., Shaughnessy, J. D., Jenkins, N. A. & Copeland, N. G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat. Genet. 12, 149–153 (1996).
Kroon, E. et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17, 3714–3725 (1998).
Thorsteinsdottir, U. et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99, 121–129 (2002).
Li, Z. et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood 121, 1422–1431 (2013).
Alcalay, M. et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood 106, 899–902 (2005).
Brunetti, L. et al. Mutant NPM1 maintains the Leukemic State through HOX expression. Cancer Cell 34, 499-512.e9 (2018).
Acknowledgements
This work was funded by a Kay Kendall Leukemia Fund Intermediate Fellowship (KKL889) awarded to RN. This work was supported by Cancer Research UK (C416/A20938) in the form of a Cancer Research UK UCL Centre PhD studentship (163011) supporting AM. Work in the Enver laboratory was supported by a Bloodwise Programme Grant (16001) and an MRC Programme Grant (MR/N000838/1). ED was funded by a Children with Cancer Project Grant awarded to RN and TE (17-250). NA was funded by a Wellcome Trust Clinical Research Training Fellowship (107387/Z/15/Z). This work was also supported by a British Heart Foundation Award (PG/12/36/29444) to GML, and JSPS KAKENHI Grant Number JP16H06276 (AdAMS) (MA and KA). We acknowledge the support of the CRUK UCL Centre FACS facility for performing cell sorting. We thank Cure Cancer @ UCL for generously providing key equipment used in this study. EG acknowledges research funding from the Barry Reed Cancer Research Fund.
Author information
Authors and Affiliations
Contributions
R.N. conceptualized the project with input from T.E. and D.L. A.M., J.K., P.D., Y.G., E.D., J.B., S.B., E.G., and R.N. performed experiments. M.A. and K.A. generated and provided the Mir142em2Card KO mice. N.A., I.J., J.K.H. and G.M.L. generated and provided tissue from Mir142HOJ2 KO mice. Mass spectrometry analysis was performed by S.B. and E.G. IDH2R140Q cDNA was provided by S.B. and A.K. RNA-seq data was processed, analyzed and archived by C.J. with input from R.N. and A.M. R.N. analyzed the data and wrote the paper with input and support from A.M., G.M., and T.E. R.N. obtained funding for the project with support from T.E. and D.L. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marshall, A., Kasturiarachchi, J., Datta, P. et al. Mir142 loss unlocks IDH2R140-dependent leukemogenesis through antagonistic regulation of HOX genes. Sci Rep 10, 19390 (2020). https://doi.org/10.1038/s41598-020-76218-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-76218-8
This article is cited by
-
miR-142 deficit in T cells during blast crisis promotes chronic myeloid leukemia immune escape
Nature Communications (2025)
-
Mutations in the miR-142 gene are not common in myeloproliferative neoplasms
Scientific Reports (2022)