Abstract
Rapeseed, a major oil crop in the world, is easily affected by low-temperature stress. A low temperature delays seed germination and increases seedling mortality, adversely affecting rapeseed growth and production. In the present study, a tolerant cultivar (Huyou21) was crossed with a susceptible genotype (3429) to develop a mapping population consisting of 574 F2 progenies and elucidate the genetic mechanisms of seed germination under low temperatures. Two quantitative trait loci (QTL) for low-temperature germination (LTG) were detected, one on chromosome A09 (named qLTGA9-1) and the other on chromosome C01 (named qLTGC1-1), using the QTL-seq approach and confirmed via linkage analysis in the mapping population. Further, qLTGA9-1 was mapped to a 341.86 kb interval between the SSR markers Nys9A212 and Nys9A215. In this region, 69 genes including six specific genes with moderate or high effect function variants were identified based on the Ningyou7 genome sequence. Meanwhile, qLTGC1-1 was mapped onto a 1.31 Mb interval between SSR markers Nys1C96 and Nys1C117. In this region, 133 genes including five specific genes with moderate effect function variants were identified. These specific genes within the two QTL could be used for further studies on cold tolerance and as targets in rapeseed breeding programs.
Similar content being viewed by others
Introduction
Rapeseed (Brassica napus L.) is a major winter crop mainly cultivated in the Yangtze River Basin of China, with a sown area of 6.6 million hectares and annual production of 13.5 million tons1,2. However, the cultivation patterns gradually changed from transplanting into no-tillage direct seeding with improvement in technology and an increase in labor cost3. The late direct seeding area under rapeseed has continuously increased in recent years, with intensive cropping development. Meanwhile, rapeseed is sensitive to temperatures below 10 °C at germination4,5. The temperature at late sowing time in China is generally below 10 °C, which leads to a low germination rate and seedling in rapeseed6, which ultimately affects yield6,7. Therefore, selecting varieties of rapeseed with a high rate of low-temperature germination (LTG) has become an important breeding goal under late direct seeding cultivation.
Different cultivars respond differently to low temperatures, and genetic factors largely control these differences. The LTG ability of crops is a complex trait controlled by quantitative trait loci (QTL)6,7,8. Several QTL mapping studies for LTG have been conducted in crops, such as rice, maize, soybean, and wheat9,10,11,12,13,14,15. Studies have found wide genotypic variation in LTG in rapeseed16,17 and associated a few QTL with low-temperature stress during seed germination and seedling stage. Xian et al.6 identified several differentially expressed genes related to low-temperature tolerance in rapeseed through transcriptome analysis. Luo et al.7 detected 22 QTL associated with low-temperature tolerance during seed germination and seedling stage through genome-wide association study (GWAS). However, genetic study on rapeseed germination under low temperature is still rare. Moreover, only a few markers associated with LTG have been developed for rapeseed breeding. Thus, it is critical to identify novel LTG QTL for fine mapping to accelerate the breeding of low-temperature tolerant varieties of rapeseed.
QTL associated with specific traits can be identified through different QTL mapping approaches, including QTL-seq. QTL-seq is an effective approach performed via bulked segregant analysis (BSA) using next-generation sequencing (NGS)18. QTL-seq has been used to rapidly identify QTL for different traits, such as spikelet fertility under heat stress in rice19, plant height in wild soybean20, and cold tolerance in wild rice21. The present study employed the F2:3 populations, derived from two inbred lines, Huyou21 (tolerant to low-temperature stress) and 3429 (susceptible to low-temperature stress), to detect the QTL related to rapeseed LTG via QTL-seq. The objectives of this study were to: (i) analyze seed germination of this population under low temperature; (ii) identify QTL for LGT from this population; and (iii) develop simple sequence repeat (SSR) markers of the LTG QTL for further fine-mapping or molecular breeding.
Results
Analysis of LTG and construction of bulks
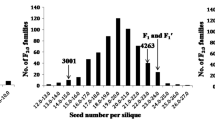
Based on a preliminary screening for LTG, the tolerant cultivar Huyou21 and the susceptible cultivar 3429 were selected for further study. At optimal temperature (20 °C), the two parents demonstrated similar germination rates (> 95%) within four days after imbibition (DAI). At low temperature (8 °C), Huyou21 exhibited excellent tolerance to cold stress, with a mean germination rate higher than 90% within four DAI. In contrast, 3429 showed high susceptibility at 8 °C, with a mean germination rate lower than 40% within four DAI (Fig. 1A). The mean germination rate of all F1:2 seeds derived from the F1 plants of 3429 × Huyou21 cross was 76.9%, and that derived from the F1′ plants of Huyou21 × 3429 cross was 70.2%. The F2 lines showed large germination rate variations, ranging from 17.5% to 100.0%, exhibiting a distribution skewed toward tolerance (Fig. 1A).
Regression analysis of parent–offspring revealed an LTG heritability estimate of 0.26 for the F2:3 families. After cold stress, 344 plants out of F2:3 families showed a germination rate larger than 90%, and 230 plants showed a germination rate equal to or less than 90%. Besides, the tolerant and susceptible plants fit well in the 9:7 ratio (χ2 = 1.58, p = 0.21) (Table S1). These results indicate the role of two dominant genes in controlling LTG in Huyou21. The F2:3 families showed pronounced variation and segregation in cold stress tolerance or sensitivity. The LTG tolerance distribution was continuous and approximately normal in the F2:3 families, indicating a quantitative inheritance of LTG. Meanwhile, the phenotypic trait indices (PTI) of F2 progeny ranged from 0.19 to 2.58 (Fig. 1B). To establish the low temperature tolerant (LT) bulk and low-temperature sensitive (LS) bulk, 30 LT and 30 LS F2 individuals were selected. The PTI of the 30 plants in the LT bulk was more than 2.33, and that of the 30 plants in the LS bulk was less than 0.99.
Resequencing and mapping of reads
The two bulks (LT bulk and LS bulk) with extreme phenotypes (the tolerant and sensitive pools) and the two parents were used to prepare the libraries for Illumina sequencing. A total of 319,059,584 reads were generated for LT bulk, 335,343,844 reads for LS bulk, 323,983,156 reads for tolerant parent (Huyou21), and 337,990,498 reads for susceptible parent (3429). Alignment of the reads to the Ningyou7 rapeseed genome sequence22 revealed 36.52 × and 37.58 × read depth and 99.17% and 99.13% coverage for LT bulk and LS bulk, respectively. Similarly, 36.41 × and 38.26 × read depth and 95.73% and 97.31% coverage were obtained for Huyou21 and 3429, respectively (Table 1). In total, 5,600,575 genome-wide SNPs and 1,215,792 InDels were obtained for LT bulk, and 5,604,534 SNPs and 1,217,972 InDels for LS bulk by comparing with the reference genome (Table S2 and Fig. S1).
Candidate genomic regions identified for LTG by QTL-seq
Further, the SNP index was calculated for each bulk to identify the candidate genomic regions related to LTG. Then, Δ(SNP-index) was calculated and plotted against the genome positions by combining the information on the SNP-index in LT bulk and LS bulk (Fig. 2). The Δ(SNP-index) was calculated by subtracting the SNP-index of LS bulk from the SNP-index of LT bulk, and plotted across the 19 rapeseed chromosomes to map the putative genomic regions associated with the phenotype for LTG. Two QTL regions on the two chromosomes A09 and C01 were detected based on Δ(SNP-index) plot (Fig. 2C), at 95% significance. The peak of qLTGA9-1 was located between 40.00 Mb and 49.73 Mb on chromosome A09 (Fig. 2D), and that of qLTGC1-1 was located between 38.90 Mb and 55.38 Mb on chromosome C01 (Fig. 2E). The results revealed one QTL related to LTG at the 9.73 Mb region of chromosome A09, named qLTGA9-1, and another QTL at the 16.48 Mb region of chromosome C01, named qLTGC1-1.
Identification of LTG QTL in F2 populations by QTL-seq. (A, B, and C) represent the SNP-index plots of LT bulk and LS bulk, and the Δ(SNP-index) plot of 19 rapeseed chromosomes from QTL-seq analysis, respectively. Δ(SNP-index) plot with statistical confidence interval under the null hypothesis of no QTL (purple, P < 0.05; orange, P < 0.01). (D)and (E) represent the significant genomic regions of 40.00–49.73 Mb on chromosome A09 and 38.90–55.38 Mb on chromosome C01 identified for LTG, respectively. The X-axis represents the chromosomes of rapeseed, and Y-axis represents the SNP-index.
Marker development and QTL fine mapping
A total of 351 SSR markers, including 163 primers in the 9.73 Mb candidate region (qLTGA9-1) on chromosome A09 and 188 primers in the 16.48 Mb candidate region (qLTGC1-1) on C01, were developed and genotyped in Huyou21 and 3429 to analyze the result of QTL-seq. Among these, 17 primer pairs (eight on A09 and nine on C01) produced steady and clear polymorphic bands between the two parents (Table 2), indicating that these primers could be used in QTL mapping for LTG. These polymorphic markers were selected for linkage map construction, and linkage analysis was performed in the 574 F2 individuals with the LTG phenotype. The genetic map and the LTG phenotypic analysis demonstrated both QTL on the predicted regions (Fig. 3). The QTL qLTGA9-1 was mapped in a 1.78 cM interval between the tightly linked markers, Nys9A212 and Nys9A215, which only explained 5.93% of the phenotypic variation for LTG but had a higher Limit of detection (LOD) value of 8.43 (Fig. 3 and Table 3). Meanwhile, qLTGC1-1 was mapped in a 16.40 cM interval between the tightly linked markers, Nys1C96 and Nys1C117, which explained a phenotypic variation of 5.39% and had a LOD value of 6.57 (Fig. 3 and Table 3).
Based on the physical position of the tightly linked markers, qLTGA9-1 was localized in a 341.86 kb interval between 44.72 Mb and 45.06 Mb on rapeseed chromosome A09, and qLTGC1-1 in the 1.31 Mb interval between 48.19 Mb and 49.50 Mb on chromosome C01. Further, annotation based on Ningyou7 genome sequence22 predicted that 69 and 133 genes were located in the qLTGA9-1 region and qLTGC1-1 region, respectively. Further, 74 SNP and 25 InDel mutations related to 32 predicted genes in the qLTGA9-1 region, and 162 SNP and 26 InDel mutations related to 35 predicted genes in the qLTGC1-1 region were screened based on the sequencing results for each QTL (Table S3 and Table 4). Of these predicted genes, only one gene in the qLTGA9-1 region was a variant (frameshift insertion/deletion) with a high effect, and five genes in qLTGA9-1 and qLTGC1-1 regions were variants (missense mutation) with moderate effects (Tables 4 and 5). These gene variants with moderate or high effects may be related to low-temperature tolerance at the generation stage in rapeseed.
The Ningyou7 annotation22 identified that seven of the predicted genes (four genes in qLTGA9-1 and three genes in qLTGC1-1) encode proteins associated with plant growth or temperature stress response as follows: ChrA09g005501 encodes a formiminotransferase, ChrA09g005507 encodes a protein phosphatase 2C (PP2C), ChrA09g005509 encodes an aminotransferase, ChrA09g005523 and ChrA09g005524 encode DRG family regulatory proteins (DFRP), ChrC01g004357 encodes a SWEET sugar transporter, ChrC01g004359 encodes a PHD-finger, and ChrC01g004400 encodes a plant invertase/pectin methylesterase inhibitor (PMEI) (Table 5).
Discussion
The late direct seeding area for rapeseed in China has continuously increased with intensive cropping development. However, low temperature occurs in late-autumn or early-winter quickly affects rapeseed germination with the delay of rapeseed seeding times6. Therefore, it is a breeding goal to select varieties with good germination under both optimal and stressed conditions to adapt to changing temperatures under late direct seeding cultivation. In the previous studies, a high degree of variability was observed in the seed germination rate among the rapeseed genotypes under low-temperature conditions. The genotypic variability helped LTG studies in rapeseed breeding to deal with cold stress under late direct-seeding conditions.
Studies have also revealed variations in LTG among plant species, which are generally affected by inheritance and environmental factors and regulated by QTL and multi-genes6,7,8. QTL mapping studies for LTG have been conducted in various crops, such as rice, maize, soybean, and wheat9,10,11,12,13,14,15; however, few QTL or genes for LTG are reported in rapeseed. QTL-seq combined with BSA and NGS is a practical approach used to identify QTL18. The method has been successfully used to rapidly identify QTL of different traits in rapeseed23,24. In this research, a segregating population was employed to detect the LTG QTL of rapeseed using QTL-seq. The analysis of the LTG in the population derived from the 3429 × Huyou21 cross revealed that two dominant nuclear genes or QTL controlled the LTG of these populations, with a heritability of 0.26. Further, QTL-seq revealed two QTL (namely qLTGA9-1 and qLTGC1-1) associated with LTG on chromosomes A09 and C01; both were verified with classical QTL analysis through map construction. Here, qLTGA9-1 was mapped between the flanking SSR markers Nys9A212 and Nys9A215, and qLTGC1-1 was mapped between Nys1C96 and Nys1C117. Besides, the study found that the QTL (qLTGA9-1) on chromosome A09 was mapped around 30.2 Mb apart from each other based on the physical position of linkage markers on ZS11 genome sequence25. Earlier, GWAS mapping by Luo et al.7 showed that the candidate genes related to seed vigor under low temperature were localized around the physical position of 3.0 Mb on chromosome A09 based on ZS11 genome sequence25. As the physical distance between the locus and the QTL identified in this study is more than 27 Mb, the QTL, including qLTGC1-1, identified are novel in rapeseed for LTG.
Furthermore, to recapitulate the physical position of the linkage markers (Nys9A212 and Nys9A215, Nys1C96 and Nys1C117) based on the Ningyou7 genome sequence22, 69 and 133 genes were predicted in qLTGA9-1 and qLTGC1-1, respectively, and 11 of these predicted genes were variants with moderate/high effect according to QTL-seq. Among these predicted genes were ChrA09g005507 in the qLTGA9-1 region encoding a PP2C and ChrC01g004357 in the qLTGC1-1 region encoding a SWEET protein. PP2C is a key player in ABA signal transduction, which plays an important role in seed germination under cold stress26,27,28, while SWEET protein is an important plant sugar transporter family and plays a crucial role in seed germination and stress response29,30. In addition, ChrA09g005523 and ChrA09g005524 in qLTGA9-1 region encode DFRPs, while ChrC01g004359 and ChrC01g004400 in qLTGC1-1 region encode a PHD-finger and a PMEI, respectively. Like PP2C or SWEET, the DFRP, PHD-finger, and PMEI proteins also play significant roles in defense response or under abiotic stress31,32,33. However, further research should analyze and functionally validate these candidate genes.
Materials and methods
Plant materials
The parents used to develop the mapping population were Huyou21 (tolerant to cold stress) and 3429 (susceptible to cold stress) (Fig. 4). Seeds of Huyou21 and 3429 cultivars were obtained from the Shanghai Academy of Agricultural Sciences. Huyou21, developed from a double-cross between rapeseed lines 9714/9711 and 84004/8920, is widely cultivated in the lower reaches of the Yangtze River Basin, China, while 3429, used as the female, is a new line derived from a self-cross plant of the hybrid variety Qinyou99. The 3429 × Huyou21 hybrids were advanced from the F1 generation by selfing to yield the F2:3 families for mapping of LTG. Besides, the seed germination of 500 seeds harvested at the same time from both parents and their generations was confirmed.
Phenotypic evaluation
Five hundred healthy and plump F3 seeds obtained per F2 plant, all together F2 seeds per F1 plant, and their parental controls were placed on two layers of filter paper with 15 mL distilled water in Petri dishes (9 cm inner diameter). The germination experiment was conducted in a low-temperature incubator set at 8 °C in a completely randomized design with three replications per treatment. The number of germinated seeds (N1) was counted on the fourth day after imbibition (DAI). Seeds that did not germinate after four days under low temperature (8 °C) were removed to normal temperature (20 °C) to start the recovery process for three days, and the number of remaining germinated seeds (N2) under normal temperature were counted to exclude the percentage of low-vigor seed influences. A seed was considered germinated once the radicle emerged and elongated 2–3 mm from the seed. The seed germination under low-temperature was calculated using the following formula:
Plants with a germination rate larger than 90% were classified as cold tolerant, and those with a germination rate equal to or less than 90% were classified as cold susceptible. The broad-sense heritability of LTG expression was estimated using parent–offspring regression methods based on the variance of parents, F1-derived F2 population, and F2-derived F3 families34. The data on germination rate were arcsine transformed to improve the homogeneity of variance35,36. Further, the phenotypic trait indices (PTI) of F2:3 families for QTL mapping of LTG were calculated using the following formula to reduce the influence of microenvironment features during cold treatments:
where X represents the grand mean of germination rate of the F2:3 family, P1 and P2 represent the germination rate of their parental controls. The PTI of F2:3 families was evaluated and analyzed via chi-square test using SAS software (v9.1, SAS Institute, Cary, NC).
Sample bulking and DNA isolation
A total of 574 F2 individuals from 3429 × Huyou21 cross were selected to build DNA bulks for QTL-seq. Thirty individuals for LT bulk and another 30 individuals for LS bulk were selected from the F2 population based on the extreme phenotype of F2:3 families for PTI under the low temperature (8 °C). Genomic DNA from LT bulk, LS bulk, F2 individuals and their parents (Huyou21 and 3429) were isolated using Plant Genomic DNA Kit (TIANGEN, China), following the instructions. DNA quality and concentration were examined by agarose gel electrophoresis (1%; w/v).
Illumina sequencing and NGS data analysis
Test-qualified genomic DNA samples from LT bulk, LS bulk, and two parents were used to construct libraries with an insert size of 350–500 bp at Shanghai OE Biotech Co., Ltd. (China) using the TruSeq DNA LT Sample Prep Kit and sequenced (150 bp pair-end reads) using an Illumina Xten platform. Raw data generated from Illumina sequencing were subjected to quality control using Trimmomatic (v0.36)37. The filtered clean reads from both parents and two DNA bulks were aligned to the rapeseed genome sequence22 using BWA software, and single nucleotide polymorphism (SNP) calling was performed with SAMtools38. The average SNP-index for each pool was calculated in 1 Mb sliding windows with a 10 kb increment. The △(SNP-index) was calculated by subtracting the SNP-index of LS bulk from that of LT bulk. All SNP-index and △(SNP-index) were calculated for all positions as previously described18,39 to identify LTG-related QTL.
SSR marker analysis and QTL fine mapping
LTG-related QTL identified by QTL-seq were validated and fine mapped through the traditional QTL mapping method40. A total of 351 SSR markers in the predicted regions were mined from the whole-genome sequence22. The SSR markers were used to survey the polymorphism between parents, which were designed with SSR Locator41 based on the parameters as previously described40. The newly developed markers were named NysX(A/C)Y markers, where Nys represents the microsatellite from the physical sequence of Ningyou7 rapeseed, the number X indicates the chromosome in subgenome (A or C), and Y represents a numerical code for the newly designed marker. Further, PCR and amplicon detection for the F2 individuals and their parents were performed as previously described42 with minor modifications. Polymorphic markers were further selected to analyze F2 population to construct a linkage map for QTL fine mapping. The genetic linkage map was drawn using the MAP functionality in QTL IciMapping v4.143, and QTL was conducted using the BIP functionality43. The map distance (cM) was calculated using the Kosambi mapping function44, and the mapping method adopted was ICIM-ADD45. The LOD threshold and recombination frequency were set at 3.0 and 0.30, respectively. The QTL were designated by the term qLTG followed by the chromosome number.
Candidate gene annotation
The candidate genes within the detected QTL for LTG were obtained based on the Ningyou7 rapeseed genome annotation22, and the functional variant effects of SNPs or Indels in each gene were predicted using SnpEff46 software.
Permission statement
All the experiments on plants, including the collection of rapeseed materials, were performed in accordance with relevant guidelines and regulations.
Conclusions
A total of 574 F2:3 families were constructed to elucidate the genetic mechanisms of seed germination under low temperature in rapeseed. Based on the QTL-seq and linkage analysis of the populations, two QTL were identified from ‘Huyou21’. One QTL was mapped to a 341.86 kb interval between the SSR markers Nys9A212 and Nys9A215 on rapeseed chromosome A09, and another was mapped to a 1.31 Mb interval between the SSR markers Nys1C96 and Nys1C117 on chromosome C01. These findings provide a basis for further studies on genetic breeding and assist in cloning candidate genes for cold tolerance in rapeseed.
Data availability
This whole genome resequencing reads used in QTL-seq has been deposited in the National Center of Biotechnology Information Sequence Read Archive (SRA) under BioProject accession number PRJNA751740.
References
Tian, Z. et al. The potential contribution of growing rapeseed in winter fallow fields across Yangtze River Basin to energy and food security in China. Resour. Conserv. Recycl. 164, 105159 (2021).
Food and Agriculture Organization of the United Nations (FAOSTAT) data sets. http://www.fao.org/faostat/en/#data/QCL
Wang, R., Cheng, T. & Hu, L. Effect of wide-narrow row arrangement and plant density on yield and radiation use efficiency of mechanized direct-seeded canola in Central China. Field Crops Res. 172, 42–52 (2015).
Kondra, Z. P., Campbell, D. C. & King, J. R. Temperature effects on germination of rapeseed (Brassica napus L. and B. campestris L.). Can. J. Plant Sci. 63, 1063–1065 (1983).
Luo, T. et al. Genome-wide association mapping unravels the genetic control of seed vigor under low-temperature conditions in rapeseed (Brassica napus L.). Plants 10, 426 (2021).
Xian, M., Luo, T., Khan, M. N., Hu, L. & Xu, Z. Identifying differentially expressed genes associated with tolerance against low temperature stress in Brassica napus through transcriptome analysis. Int. J. Agric. Biol. 19, 273–281 (2017).
Wang, X. et al. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 69, 413–421 (2018).
Chinnusamy, V., Zhu, J. K. & Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55 (2010).
Baga, M. et al. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct. Integr. Genomics 7, 53–68 (2007).
Zhang, W. et al. Genetic overlap of QTL associated with low-temperature tolerance at germination and seedling stage using BILs in soybean. Can. J. Plant Sci. 92, 1381–1388 (2012).
Hu, S., Lubberstedt, T., Zhao, G. & Lee, M. QTL mapping of low-temperature germination ability in the maize IBM Syn4 RIL population. PLoS ONE 11, e0152795 (2016).
Jiang, N. et al. Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice. Front. Plant Sci. 8, 1223 (2017).
Li, X. et al. QTL mapping in three connected populations reveals a set of consensus genomic regions for low temperature germination ability in Zea mays L. Front. Plant Sci. 9, 65 (2018).
Thapa, R., Tabien, R. E., Thomson, M. J. & Septiningsih, E. M. Genome-wide association mapping to identify genetic loci for cold tolerance and cold recovery during germination in rice. Front. Genet. 11, 22 (2020).
Pei, R. et al. Mapping QTLs controlling low-temperature germinability in rice by using single segment substitution lines derived from 4 AA-genome species of wild rice. Euphytica 217, 58 (2021).
Russo, V. M., Bruton, B. D. & Sams, C. E. Classification of temperature response in germination of Brassicas. Ind. Crops Prod. 31, 48–51 (2010).
Zhang, C. et al. Evaluation of the low-temperature tolerance of rapeseed genotypes at the germination and seedling emergence stages. Crop Sci. 59, 1709–1717 (2019).
Takagi, H. et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183 (2013).
Nubankoh, P. et al. QTL-seq reveals genomic regions associated with spikelet fertility in response to a high temperature in rice (Oryza sativa L.). Plant Cell Rep. 39, 149–162 (2020).
Zhang, X. et al. Combining QTL-seq and linkage mapping to fine map a wild soybean allele characteristic of greater plant height. BMC Genomics 19, 226 (2018).
Luo, X. et al. Rapid mapping of candidate genes for cold tolerance in Oryza rufipogon Griff. by QTL-seq of seedlings. J. Integr. Agric. 17, 265–275 (2018).
Zou, J. et al. Genome-wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed. Plant Biotechnol. J. 17, 1998–2010 (2019).
Wang, H. et al. Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-seq. Sci. Rep. 6, 38493 (2016).
Tudor, E. H. et al. QTL-seq identifies BnaFT.A02 and BnaFLC.A02 as candidates for variation in vernalization requirement and response in winter oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2466–2481 (2020).
Sun, F. et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468 (2017).
Nishimura, N. et al. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50(6), 935–949 (2007).
Hu, X. et al. Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. J. Plant Physiol. 167, 1307–1315 (2010).
Endo, A. et al. Ectopic expression of mutated type 2C protein phosphatase OsABI-LIKE2 decreases abscisic acid sensitivity in Arabidopsis and rice. Sci. Rep. 8, 12320 (2018).
Klemens, P. et al. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 163, 1338–1352 (2013).
Gautam, T. et al. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 46, 2327–2353 (2019).
O’Connell, A., Robin, G., Kobe, B. & Botella, J. R. Biochemical characterization of Arabidopsis developmentally regulated G-proteins (DRGs). Protein Expres. Purif. 67, 88–95 (2009).
Rocchi, V. et al. Intron retention regulates the expression of pectin methyl esterase inhibitor (Pmei) genes during wheat growth and development. Plant Biol. 14, 365–373 (2012).
Miura, K., Na, R. & Suzaki, T. The PHD finger of Arabidopsis SIZ1 recognizes trimethylated histone H3K4 mediating SIZ1 function and abiotic stress response. Commun. Biol. 3, 23 (2020).
Nyquist, W. E. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 10, 235–322 (1991).
Badger, K. S. & Ungar, I. A. The effects of salinity and temperature on the germination of the inland halophyte Hordeum jubatum. Can. J. Bot. 67, 1420–1425 (1989).
Gorai, M., Gasmi, H. & Neffati, M. Factors influencing seed germination of medicinal plant Salvia aegyptiaca L. (Lamiaceae). Saudi. J. Biol. Sci. 18, 255–260 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. et al. 1000 Genome project data processing subgroup: The sequence alignment/map format and SAMtools. Bioinformatics 25(16), 1754–1760 (2009).
Abe, A. et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178 (2012).
Zhu, J. F. et al. QTL and candidate genes associated with common bacterial blight resistance in the common bean cultivar Longyundou 5 from China. Crop J. 4, 344–352 (2016).
Maia, L. C. et al. SSR locator: tool for simple sequence repeat discovery integrated with primer design and PCR simulation. Int. J. Plant Genomics 2008, 1–9 (2008).
Zhu, J. F. et al. Development of genome-wide SSR markers in rapeseed by next generation sequencing. Gene 798, 145798 (2021).
Lei, M., Li, H. H., Zhang, L. Y. & Wang, J. K. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 121, 269–283 (2015).
Kosambi, D. D. The estimation of map distances from recombination values. Ann. Hum. Genet. 12, 172–175 (1943).
Li, H. H., Ye, G. Y. & Wang, J. K. A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374 (2007).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff, Fly 6(2), 80–92 (2012).
Acknowledgements
The authors would like to thank all the reviewers who participated in the review and MJEditor for its linguistic assistance during the preparation of this manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant No. 31901502).
Author information
Authors and Affiliations
Contributions
J.Z. designed and performed the experiments, collected and conducted the data analysis, and wrote the manuscript. X.Z. provided the genetic materials and assisted in phenotype investigation. W.W., M.J. and L.Y. worked on the materials sowing. J.Z. and X.Z. conceived the study and finalized the manuscript. All authors read and approved the final version of paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, J., Wang, W., Jiang, M. et al. QTL mapping for low temperature germination in rapeseed. Sci Rep 11, 23382 (2021). https://doi.org/10.1038/s41598-021-02912-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-021-02912-w
This article is cited by
-
Jasmonic Acid: A Key Elicitor of Cold Stress Tolerance in Horticultural Crops
Journal of Plant Growth Regulation (2025)
-
QTL mapping for seed density per silique in Brassica napus
Scientific Reports (2023)