Abstract
Genome-wide association studies in Europeans and Asians have identified numerous variants in the transmembrane protease serine 6 (TMPRSS6) and transferrin (TF) genes that are associated with changes in iron status. We sought to investigate the effects of common TMPRSS6 and TF gene SNPs on iron status indicators in a cohort of healthy Africans from rural Gambia. We measured iron biomarkers and haematology traits on individuals participating in the Keneba Biobank with genotype data on TMPRSS6 (rs2235321, rs855791, rs4820268, rs2235324, rs2413450 and rs5756506) and TF (rs3811647 and rs1799852), n = 1316. After controlling for inflammation, age and sex, we analysed the effects of carrying either single or multiple iron-lowering alleles on iron status. TMPRSS6 rs2235321 significantly affected plasma hepcidin concentrations (AA genotypes having lower hepcidin levels; F ratio 3.7, P = 0.014) with greater impact in individuals with low haemoglobin or ferritin. No other TMPRSS6 variant affected hepcidin. None of the TMPRSS6 variants nor a TMPRSS6 allele risk score affected other iron biomarkers or haematological traits. TF rs3811647 AA carriers had 21% higher transferrin (F ratio 16.0, P < 0.0001), 24% higher unsaturated iron-binding capacity (F ratio 12.8, P < 0.0001) and 25% lower transferrin saturation (F ratio 4.3, P < 0.0001) compared to GG carriers. TF rs3811647 was strongly associated with transferrin, unsaturated iron-binding capacity (UIBC) and transferrin saturation (TSAT) with a single allele effect of 8–12%. There was no association between either TF SNP and any haematological traits or iron biomarkers. We identified meaningful associations between TMPRSS6 rs2235321 and hepcidin and replicated the previous findings on the effects of TF rs3811647 on transferrin and iron binding capacity. However, the effects are subtle and contribute little to population variance. Further genetic and functional studies, including polymorphisms frequent in Africa populations, are needed to identify markers for genetically stratified approaches to prevention or treatment of iron deficiency anaemia.
Similar content being viewed by others
Introduction
The discovery of hepcidin and the molecular mechanisms modulating its relationship with iron metabolism have brought new insights into how iron is regulated in the human body1,2. Subsequently, several genome-wide studies (GWASs) have revealed single nucleotide polymorphism (SNPs) in genes involved in hepcidin regulatory pathways, that are associated with impaired iron status3,4,5. The most common SNPs associated with low iron status are in the TMPRSS6 gene, encoding the matriptase-2 protein6,7,8. TMPRSS6 suppresses hepcidin synthesis, and its impaired function has been associated with inappropriately high hepcidin, which restricts iron absorption by the duodenum and iron mobilisation from storage6,9,10. Impaired TMPRSS6 activity has been implicated in the development of iron-refractory iron deficiency anaemia (IRIDA)8.
So far, more than 50 SNPs within the TMPRSS6 gene have been reported to be associated with impaired iron status. The most commonly reported SNPs are rs855791 and rs4820268 and rs22353213,4,11,12,13,14. However, most studies linking TMPRSS6 SNPs and low iron status were conducted in Europeans and Asians. Genetic variations in the TMPRSS6 gene has been linked to variations in iron status indicators in different populations across the world, including in India12, Turkey15 and Australia16.
Also, SNPs in the transferrin (TF) gene have been associated with altered iron status17,18,19. The most common TF SNP to be associated with the risk of iron deficiency is rs381164717,20,21,22. This SNP is associated with low iron status in different populations globally, including in African populations23. However, little information exists on the effect TF SNPs on low iron status, particularly in settings with high anaemia burden.
Despite efforts to identify genetic risk factors for anaemia, very few such studies have been reported from sub-Saharan Africa24. To the best of our knowledge, no study has been done in West Africa to assess the effects of genetic variants in hepcidin and iron regulatory genes on low iron status. This is particularly important given that West Africa is one of the regions with the highest prevalence of anaemia25. In this study, we investigated the association between common SNPs in the TMPRSS6 and TF genes, and iron indicators in healthy individuals from the rural Gambia.
Results
Baseline characteristics of the study population are presented in Table 1. Due mostly to out-migration of males there was a slight sex bias (54.2% were female). There were significant differences between the sexes in age, RBC count and RBC indices, serum iron and transferrin.
TMPRSS6 variants
All the SNPs investigated were in Hardy–Weinberg Equilibrium. Also, all were in low linkage disequilibrium (LD) in this study population, except rs4820268 and rs2413450 which have r2 = 0.7 (Fig. 1). Among the SNPs we investigated, TMPRSS6 rs2235324 had the highest minor allele frequency (MAF) in our study population (45%), and TMPRSS6 rs855791 and TF rs1799852 had the lowest MAF (7% each)24.
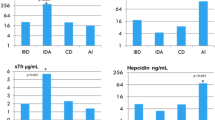
There was no detectable influence of sex on any associations, so sex was discarded from the models. None of the TMPRSS6 SNPs studied showed any association with any of the iron status markers (ferritin, serum iron, transferrin, TSAT, sTfR, TIBC or UIBC) or haematological variables (Hb, MCV, MCH, MCHC, RDW, RBC or Hct) or with CRP. However, hepcidin levels varied significantly by rs2235321 genotype (Fig. 2A) with lower hepcidin in the AA homozygotes, 19% than GG carriers (F ratio 3.70, P = 0.014). Note that Bonferroni correction for having analysed 6 SNPs would render the rs2235321 of marginal significance. These trends were stronger in subjects with lower Hb (Fig. 2B) and lower ferritin levels (Figs. 2C). The other SNPs had no detectable influence on hepcidin.
The effects of TMPRSS6 rs2235321 on plasma hepcidin levels. (A) All data (GG n = 416, GA n = 586, AA n = 262). ANOVA P for trend = 0.004. (B) Sample divided into high and low Hb (< 11.5 g/dl). High Hb (GG n = 245, GA n = 344, AA n = 162). ANOVA P for trend = 0.02. Low Hb (GG n = 171, GA n = 242, AA n = 100). ANOVA P for trend = 0.0002. (C) Sample divided above and below median ferritin (< 26 ng/ml). High ferritin (GG n = 247, GA n = 313, AA n = 155). ANOVA P for trend = 0.0004. Low ferritin (GG n = 169, GA n = 273, AA n = 107). ANOVA P for trend = NS.
Despite the lack of significant association with 5 of the 6 SNPs, we investigated whether allele risk score (ARS) was a significant predictor of hepcidin by using published data on the direction of association to allocate a score of 0, 1 or 2 to allele combinations for each SNP and summing the scores across all SNPs (Fig. 3). ANOVA across all ARS revealed no association with plasma hepcidin (F-ratio = 1.01, P = 0.458).
TF variants
The two TF SNPs were also tested in combination and individually. Rs1799852 showed no association with any outcomes. However, rs3811647 was strongly associated with transferrin levels (Fig. 4A) with or without the inclusion of sex and age as co-variables (F ratio 16.0, P < 0.0001). There was an apparent allele dose effect with AA homozygotes having 21% higher transferrin than GG. TIBC (partially computed from transferrin) was similarly affected with a 16% higher value for the AA genotype (F ratio 14.0, P < 0.0001).
The effects of TF rs3811647 and rs1799852 on plasma iron binding capacity and TSAT. (A) Transferrin rs3811647 All data (GG n = 720, GA n = 215, AA n = 24). ANOVA P for trend < 0.0001. Sample divided above and below median ferritin (< 26.9 ng/ml). High ferritin (GG n = 360, GA n = 108, AA n = 15). ANOVA P for trend < 0.0001. Low ferritin (GG n = 360, GA n = 107, AA n = 11). ANOVA P for trend ≤ 0.0001. rs1799852 All data (GG n = 839, GA n = 116, AA n = 4). ANOVA P for trend = NS. (B) UIBC rs3811647 All data (GG n = 985, GA n = 301, AA n = 28). ANOVA P for trend < 0.0001. Sample divided above and below median ferritin (< 26 ng/ml). High ferritin (GG n = 474, GA n = 147, AA n = 16). ANOVA P for trend < 0.0001. Low ferritin (GG n = 511, GA n = 154, AA n = 12). ANOVA P for trend < 0.0001. rs1799852 All data (GG n = 1139, GA n = 116, AA n = 9). ANOVA P for trend = NS. (C) TSAT rs3811647 All data (GG n = 720, GA n = 215, AA n = 24). ANOVA P for trend < 0.0001. Sample divided above and below median ferritin (< 26 ng/ml). High ferritin (GG n = 360, GA n = 108, AA n = 15).
UIBC, which is directly measured rather than computed, showed the same pattern (Fig. 4B) with 24% higher values in individuals carrying AA (F ratio 12.8, P < 0.0001). Serum iron was not significantly associated with the rs3811647 genotype. So, on account of the raised TIBC and transferrin, TSAT was lower in the AA group (by 25%, with a single allele effect of 12.5%) (F ratio 4.3, P < 0.0001) (Fig. 4C). The transferrin, UIBC and TSAT associations were robust when we separated the subjects into above and below the median ferritin value (Fig. 4A–C). None of the other iron markers was affected nor was there any influence on any of the haematological markers, hepcidin or CRP.
Discussion
Based upon prior GWAS studies, pathway analysis, availability on the Illumina Exome Array, and having a high minor allele frequency in our Gambian population we studied the effect of six candidate SNPs in TMPRSS6 and two in TF on multiple indices of iron and haematological status in 1316 individuals from 1 to 87 years age, from the Keneba Biobank at the MRCG @ LSHTM, in the Gambia. We found weak evidence that one TMPRSS6 SNP (rs2235321) had lower hepcidin levels in the variant (AA) homozygotes with an indication of an allele dose effect. One TF variant (rs3811647) showed increased transferrin, TIBC and UIBC levels in the AA homozygotes and lower levels of TSAT.
In this population, none of the other variants was significantly associated with any of the iron, haematological, hepcidin or inflammation markers. Applying an allele risk score approach to the 6 TMPRSS6 variants also yielded no detectable association with any outcomes. Most previous research on the effects of our candidate SNPs were conducted in non-African populations, and there is no prior data on West Africans. The associations we observed differ from results obtained in other populations. TMPRSS6 rs855791 is a non-synonymous SNP that has been widely reported to influence iron parameters and to be associated with the risk of IDA in Europeans14,19 and Asians12,26. We found no such associations between rs855791 and iron status biomarkers.
The TMPRSS6 rs2235321 is a synonymous variant which has been reported to associate with benign microcytic anaemia27. We did not find any other reported associations. Our data confirm a null effect on hepcidin even in a population with high levels of anaemia and low iron status. A meta-analysis of GWAS on the genetic determinants of hepcidin did not identify any TMPRSS6 SNP that is significantly associated with hepcidin concentration13. It is important to notice that our candidate gene approach demonstrated a relatively small effect of TMPRSS6 rs2235321 on hepcidin (single allele effect of 9.5%), and this needs to be considered when designing GWAS.
TF rs3811647 is an intron variant on the transferrin gene with extensive prior evidence for functional effects. In discovery and replication GWAS analyses of cohorts from Italy and the USA, Pichler et al.28 confirmed the association between rs3811647 and transferrin levels. In a subsequent GWAS analysis, Mclaren et al. showed that TF rs3811647 is associated with serum TIBC19. Also, Blanco-Rojo et al.21 demonstrated that rs3811647 influenced transferrin gene expression in liver. Previously, Benyamin et al. showed that three variants in TF (rs3811647, rs1799852 and rs2280673) plus the HFE C282Y mutation explained ∼40% of genetic variation in serum transferrin (p = 7.8 × 10−25)17. Our data are suggestive of an allele dose–response relationship with a single allele effect of 9.5% for transferrin levels (higher with the A variant) and a reverse effect of about 12.5% for TSAT.
Other investigators have reported associations between the TF rs1799852 and iron status. In a study of female black South Africans, Gichohi et al. reported that heterozygotes at TF rs1799852 (AG) had lower iron status (low serum ferritin and body iron, and higher sTfR concentrations) than the homozygotes (AA)23. This suggested that rs1799852 AA might be protective against low iron status. Similarly, Benyamin et al. reported that the TF rs1799852 was associated with lower transferrin concentration and the risk of haemochromatosis29. Furthermore, Blanco-Rojo et al.18 reported that the TF rs1799852 A allele was associated with low serum transferrin concentration, and it compensated for the effect of rs3811647 A allele on the risk of IDA. The authors further suggested that carrying TF rs3811467 G allele simultaneously with rs1799852 A allele and HFE C282Y and H63D might be protective against low iron status as they increase the susceptibility to iron overload18. In the present study, we could not include the HFE C282Y and H63D variants, because their MAF in Gambians is extremely low (0.4% and 0% respectively)30. Our failure to replicate the prior findings for rs1799852 may be ascribed to the fact that we had only nine individuals homozygous for the A allele. However, in contrast to the South African data23, we also found no evidence for differences in any of the iron markers between rs1799852 GG (n = 854) and AG (n = 117).
This study has strengths and weaknesses. We used highly standardised laboratory assays to measure seven markers of iron status, seven haematological traits, plus hepcidin and CRP. The sample size was large in the context of candidate gene studies, and we spanned the age range 1–87 years (with appropriate adjustment for age and sex in the analyses). The population generally has marginal iron status and high levels of anaemia which might better expose underlying genetic effects. This may suggest that the impact of underlying genetic effects on iron status may be amplified in individuals with low iron status.
One of the limitations of the study is that we had only six TMPRSS6 and two TF SNPs available on the exome chip. By definition, these SNPs had been curated onto the chip because of prior evidence of functionality. Another limitation is that for some of the SNPs (notably rs1799852 discussed above) we had very few individuals homozygous for the variant allele. In conducting this study, our initial objective was to explore whether common genetic variants in iron regulatory pathways might have a significant influence on the risk of iron deficiency and iron-deficiency anaemia in African populations. Despite selecting the variants with some strong prior evidence of functionality, our data did not reveal significant genetic effects that on the risk of anaemia to support stratified medicine approach in African population. Even where we observed associations (effects of TMPRSS6 rs2235321 on plasma hepcidin, and TF rs3811647 on iron transporting capacity and TSAT), the single allele effect sizes approximated 10%, and there were few people homozygous for the variant allele. Furthermore, there may be inherent compensatory mechanisms because none of the variants had any effect on other markers of iron or haematological status.
In this study, we identified a modest association between TMPRSS6 rs2235321 and hepcidin and replicated the previous findings on the effects of TF rs3811647 on serum transferrin. However, the overall population attributable risk conferred by these known genetic factors is negligible. As these variants were mainly studied in European and Asian populations, it is possible that other genetic variants in these genes will be more informative for iron studies in African populations, and this needs to be addressed to develop genetically stratified approaches to prevention or treatment of iron deficiency anemia.
Methods
Study populations and sample collection
This study utilised a cohort of healthy individuals, enrolled in the Keneba Biobank at MRCG@LSHTM31. Based on the availability of genotype data, we studied 1316 individuals aged 1–87 years (54.2% females). Each participant was interviewed and had a basic health examination, and those with significant health conditions excluded. After an overnight fast, a venous blood sample was collected in EDTA and lithium heparin tubes. DNA was extracted from cell pellets using standard procedures and stored at − 70 °C. Plasma samples from lithium heparin anticoagulant were stored at − 70 °C freezers until analysis.
Haematology and iron biomarker measurements
Full blood count (FBC) was performed within 4 h of sample collection using a Medonic M-Series automated haematology analyser (Boule Medical, Sweden) and results analysed for haemoglobin (Hb), mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH), mean corpuscular Hb concentration (MCHC), red cell distribution width (RDW), red blood cell number (RBC) and haematocrit (Hct). Iron biomarkers [serum iron, transferrin, ferritin, unsaturated iron-binding capacity (UIBC), soluble transferrin receptor (sTfR) and the inflammation marker C-reactive protein (CRP)] were measured using an automated biochemistry analyser (COBAS Integra 400 Plus, Roche Diagnostics). Total iron-binding capacity (TIBC) and transferrin saturation (TSAT) were calculated from UIBC and plasma iron (TIBC = plasma iron + UIBC) and TSAT = [plasma iron/TIBC] × 100). Plasma hepcidin was quantified using competitive ELISA (Bachem Hepcidin-25; Penninsula Laboratories International).
Genotyping
This study population was genotyped using the Illumina Infinium 240 K Human Exome Beadchip (v1.0 and v1.1), as previously described24, in which 848 SNPs were genotyped. Genotype calling was performed using data-driven clustering (Genome Studio, Illumina, CA, USA). The TMPRSS6 rs2235321, rs855791, rs4820268, rs2235324, rs2413450 and rs5756506, and TF rs3811647 and rs1799852 were selected for inclusion in this study based on their previously published association with iron status.
Genotype combinations and allele risk scores
For both the TMPRSS6 and TF SNPs, we generated genotype combinations and allele risk scores (ARS) by summing up the genotypes and the number of risk alleles respectively, from all the SNPs an individual carried. Risk alleles were defined as the alleles that are previously reported to be associated with iron-lowering at each SNP. For each SNP, genotypes were assigned 0, 1 or 2, with risk alleles assigned 1 and the alternate allele assigned 0. Thus, homozygous for the risk allele scored 2 and homozygous for non-risk alleles were scored 0, Table S1 (TMPRSS6 SNPs) and Table S2 (TF SNPs).
A total of 94 genotype combinations from the six TMPRSS6 SNPs were found in our population (Table S3). We investigated the effects of individual TMPRSS6 and TF SNPs on all the iron biomarkers and haematology phenotypes. In addition, based on the functional role of TMPRSS6 on hepcidin regulation, and its effects on iron status, we investigated the effects of TMPRSS6 ARS on hepcidin (Table S4). Similarly, we assessed the effects of TF SNPs’ genotype combinations and TF ARS on transferrin level.
Statistical analysis
The effects of genetic variants (genotypes of single SNPs or combinations of multiple SNPs) on iron biomarkers were determined by linear modelling with iron and haematological traits as response variables and genotype as dependent variables. Age, sex, inflammation (CRP) were added as covariates where indicated. We tested the effects each SNP individually and in combinations on iron biomarkers. Hepcidin, ferritin and CRP were log transformed. We added log ferritin as a covariate when analysing the effects of genotype on plasma hepcidin because TMPRSS6 modulates the interaction between iron status and hepcidin gene expression. Furthermore, we stratified the study population based on haemoglobin and ferritin levels and determined the effects of genotype on each sub-population. For each sub-population, we used analysis of variance (ANOVA) to determine the effects of individual SNPs on iron biomarkers. Bonferroni correction was applied to account for multiple testing. The statistical analyses were conducted using R statistical software32 and DataDesk Version 7.0.2 (Data Description Inc, Ithaca).
Ethics
The study was approved by the MRCG@LSHTM/Gambia Government Ethics Committee (SCC1185). A written informed consent was obtained from each study participant at the age of 18 years and above. For participants under the age of 18 years, a written informed consent was obtained from a parent and/or a legal guardian. All experiments were conducted in accordance with the MRCG at LSHTM and Gambia Government Ethics Committee guidelines and in accordance with the guidelines of the Helsinki Declaration.
Consent for publication
The consent for publication has been obtained from all the participants.
Data availability
The datasets generated and/or analysed during the current study are available upon written request sent to the MRCG at LSHTM/Gambia Government Joint Ethics Committee.
Abbreviations
- GWAS:
-
Genome-wide association studies
- SNPs:
-
Single nucleotide polymorphisms
- IRIDA:
-
Iron-refractory iron deficiency anaemia
- TMPRSS6 :
-
Transmembrane protease serine 6
- TF :
-
Transferrin
- HFE:
-
Haemochromatosis factor
- ARS:
-
Allele risk score
- FBC:
-
Full blood count
- RBC:
-
Red blood cells
- TSAT:
-
Transferrin saturation
- sTfR:
-
Soluble transferrin receptor
- UIBC:
-
Unsaturated iron binding capacity
- TIBC:
-
Total iron binding capacity
- CRP:
-
C-reactive protein
- LD:
-
Linkage disequilibrium
- EDTA:
-
Ethylenedimethyltetraacetic acid
- DNA:
-
Deoxyribonucleic acid
- MCV:
-
Mean corpuscular volume
- HCT:
-
Haematocrit
- MCH:
-
Mean corpuscular haemoglobin
- MCHC:
-
Mean corpuscular haemoglobin concentration
- ELISA:
-
Enzyme-linked immunosorbent assay
- MRCG at LSHTM:
-
Medical Research Council Unit the Gambia at London School of Hygiene & Tropical Medicine
- NFKBIL1:
-
Nuclear factor kB inhibitor-like protein 1
References
Ganz, T. Systemic iron homeostasis. Physiol. Rev. 93(4), 1721–1741 (2013).
Mleczko-Sanecka, K. et al. Unbiased RNAi screen for hepcidin regulators links hepcidin suppression to proliferative Ras/RAF and nutrient-dependent mTOR signaling. Blood 123(10), 1574–1585 (2014).
Benyamin, B. et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat. Genet. 41(11), 1173–1175 (2009).
Chambers, J. C. et al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat. Genet. 41(11), 1170–1172 (2009).
Li, J. et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 22(7), 1457–1464 (2013).
Lee, P. Role of matriptase-2 (TMPRSS6) in iron metabolism. Acta Haematol. 122(2–3), 87–96 (2009).
Heeney, M. M. & Finberg, K. E. Iron-refractory iron deficiency anemia (IRIDA). Hematol. Oncol. Clin. N. Am. 28(4), 637–652 (2014).
Finberg, K. E. et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat. Genet. 40(5), 569–571 (2008).
De Falco, L. et al. Iron refractory iron deficiency anemia. Haematologica 98(6), 845–853 (2013).
Ramsay, A. J., Hooper, J. D., Folgueras, A. R., Velasco, G. & Lopez-Otin, C. Matriptase-2 (TMPRSS6): A proteolytic regulator of iron homeostasis. Haematologica 94(6), 840–849 (2009).
Pei, S.-N. et al. TMPRSS6 rs855791 polymorphism influences the susceptibility to iron deficiency anemia in women at reproductive age. Int. J. Med. Sci. 11(6), 614–619 (2014).
Bhatia, P., Singh, A., Hegde, A., Jain, R. & Bansal, D. Systematic evaluation of paediatric cohort with iron refractory iron deficiency anaemia (IRIDA) phenotype reveals multiple TMPRSS6 gene variations. Br. J. Haematol. 177(2), 311–318 (2017).
Galesloot, T. E. et al. Associations of common variants in HFE and TMPRSS6 with iron parameters are independent of serum hepcidin in a general population: A replication study. J. Med. Genet. 50(9), 593–598 (2013).
Delbini, P. et al. Genetic variability of TMPRSS6 and its association with iron deficiency anaemia: Short report. Br. J. Haematol. 151(3), 281–284 (2010).
Batar, B. et al. The role of TMPRSS6 gene variants in iron-related hematological parameters in Turkish patients with iron deficiency anemia. Gene 673, 201–205 (2018).
Ji, Y., Flower, R., Hyland, C., Saiepour, N. & Faddy, H. Genetic factors associated with iron storage in Australian blood donors. Blood. https://doi.org/10.2450/2016.0138-16 (2018).
Benyamin, B. et al. Variants in TF and HFE explain ∼40% of genetic variation in serum-transferrin levels. Am. J. Hum. Genet. 84(1), 60–65 (2009).
Blanco-Rojo, R. et al. Four variants in transferrin and HFE genes as potential markers of iron deficiency anaemia risk: An association study in menstruating women. Nutr. Metab. 8(1), 69 (2011).
McLaren, C. E. et al. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS ONE 7(6), e38339 (2012).
Manjari, K. S. et al. Transferrin (rs3811647) gene polymorphism in iron deficiency anemia. Mol. Cytogenet. 7(Suppl 1), P38 (2014).
Blanco-Rojo, R., Bayele, H. K., Srai, S. K. S. & Vaquero, M. P. Intronic SNP rs3811647 of the human transferrin gene modulates its expression in hepatoma cells. Nutr. Hosp. 27(6), 2142–2145 (2012).
McLaren, C. E. et al. Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS ONE 6(3), e17390 (2011).
Gichohi-Wainaina, W. N. et al. Common variants and haplotypes in the TF, TNF-α, and TMPRSS6 genes are associated with iron status in a female black south African population. J. Nutr. 145(5), 945–953 (2015).
Jallow, M. W., Cerami, C., Clark, T. G., Prentice, A. M. & Campino, S. Differences in the frequency of genetic variants associated with iron imbalance among global populations. PLoS ONE 15(7), e0235141 (2020).
Kassebaum, N. J. The global burden of anemia. Hematol. Oncol. Clin. N. Am. 30(2), 247–308 (2016).
An, P. et al. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum. Mol. Genet. 21(9), 2124–2131 (2012).
National Center for Biotechnology Information. NM_153609.3(TMPRSS6):c.2217C>T (p.Tyr739=). ClinVar Genomic Variation as It Relates to Human Health. (2020) (accessed 15 August 2020); https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000262726.2.
Pichler, I. et al. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum. Mol. Genet. 20(6), 1232–1240 (2011).
Benyamin, B. et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 5(1), 4926 (2014).
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526(7571), 68–74 (2015).
Hennig, B. J. et al. Cohort Profile: The Kiang West Longitudinal Population Study (KWLPS)—A platform for integrated research and health care provision in rural Gambia. Int. J. Epidemiol. https://doi.org/10.1093/ije/dyv206 (2017).
R Core Team. A Language and Environment for Statistical Computing (2018).
Acknowledgements
The authors wish to thank Ebrima A. Sise, Alhassan Colley, Ebrima Bah for assisting in the laboratory analysis, Kabiru Ceesay and the Biobank Field Team for coordinating the participant recruitment for the Biobank Project; Dr Branwen Hennig for setting up the Keneba Biobank and for her expert advice; the Keneba Data Management Team for all of their help.
Funding
This research was funded by the UK Medical Research Council (Grant MC-A760-5QX00) and the UK Department for International Development under the MRC-DFID Concordat agreement. The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
M.W.J., A.M.P., S.C. and C.C. designed research; M.W.J., S.C. and C.C. conducted the research; M.W.J. analyzed the data with input from C.C., A.M.P. and S.C.; M.W.J. wrote the paper with input from all authors; C.C. and M.W.J. had primary responsibility for final content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jallow, M.W., Campino, S., Prentice, A.M. et al. Association of common TMPRSS6 and TF gene variants with hepcidin and iron status in healthy rural Gambians. Sci Rep 11, 8075 (2021). https://doi.org/10.1038/s41598-021-87565-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-021-87565-5