Abstract
Ectopic pregnancy (EP) is associated with high maternal morbidity and mortality. Ultrasonography is the only dependable diagnostic tool for confirming an ectopic pregnancy. In view of inadequate early detection methods, women suffer from a high-life risk due to the severity of EP. Early detection of EP using pathological/molecular markers will possibly improve clinical diagnosis and patient management. Salivary proteins contain potential biomarkers for diagnosing and detecting various physiological and/or pathological conditions. Therefore, the present investigation was designed to explore the salivary proteome with special reference to EP. Gel-based protein separation was performed on saliva, followed by identification of proteins using Liquid Chromatography-Tandem Mass Spectrometry (LC–MS/MS). Totally, 326 proteins were identified in the salivary samples, among which 101 were found to be specific for ruptured ectopic pregnancy (EPR). Reactome analysis revealed innate immune system, neutrophil degranulation, cell surface interactions at the vascular wall, and FCERI-mediated NF-kB activation as the major pathways to which the salivary proteins identified during EPR are associated. Glutathione-S-transferase omega-1 (GSTO1) is specific for EPR and has been reported as a candidate biomarker in the serum of EPR patients. Therefore, saliva would be a potential source of diagnostic non-invasive protein biomarker(s) for EP. Intensive investigation on the salivary proteins specific to EP can potentially lead to setting up of a panel of candidate biomarkers and developing a non-invasive protein-based diagnostic kit.
Similar content being viewed by others
Introduction
Ectopic pregnancy or extrauterine pregnancy (EP) is a condition in which the implantation and development of the fertilized ovum occurs outside to the uterus. In most of the EP cases implantation occurs in the fallopian tube, which is referred to as tubal pregnancy. In some cases non-tubal ectopic pregnancy occurs in which the implantation occurs in the ampulla, ovary, cervix, peritoneal cavity, etc.1. Moreover, EP is considered as a serious problem since it is known to lead to maternal mortality and morbidity2,3,4. The highest incidence of EP has been recorded in the African continent5. In America the rate of EP has been increasing dramatically over the past decade6. In India the incidence of maternal death due to ectopic pregnancy, in a 6 year retractive study, was 3.5–7%7. The incidence is more common in women aged 26–30 years8. According to a recent report there is increased incidence of EP due to COVID-19 infection9, the reason for which is that it could not be diagnosed based on the symptoms of abdominal pain (high blood flow in abdomen) and/or vaginal bleeding. The clinical diagnosis of EP starts with measurement of serum βhCG and progesterone, but ultrasound imaging is possibly the reliable diagnostic method. Depending on the severity of the condition, methotrexate treatment or surgical procedure is the possible clinical management practice for EP patients.
Human saliva plays a vital role in food ingestion, digestion, and oral health. Saliva is one of the few biological fluids in humans that offers a potential non-invasive source of biomolecules to represent the physiological statuses and disease conditions. Saliva contains many electrolytes, proteins (i.e., mucins, enzymes, and immunoglobulins), lipids, hormones, and other molecules10. Human saliva, put together, contains more than 3652 proteins and 12,562 peptides, accounting for 51% of proteins, and 79% of plasma peptides, respectively11,12. The salivary proteome analysis reveals that amylase and mucin constitute a major portion of the proteins13, but the concentration varies from person to person14.
Over the past decade, exploration of body fluid proteins has been considered as one of the plausible approaches to find candidate biomarkers for various diseases in the humans15,16,17,18. Thus, salivary proteins have been recommended as biomarkers to predict disease conditions such as Sjogren’s syndrome19, lung cancer20, oral cancer21, several systemic diseases22, HIV infection23,24, dental pellicle development25, hyperglycemia26, etc. Recently, it has been reported that salivary hormones and proteins have the potential to predict ovulation in women27,28. Saliva offers advantages such as easy non-invasive sampling, potential for large-scale studies, and the development of point-of-care platforms. Therefore, efforts are being put to compare saliva and serum/plasma composition for disease monitoring29. Additionally, salivary proteome analysis has provided valuable insights into the pathology of Type 1 diabetes in a pediatric population30, served as a fluid signature for inflammatory and immune-mediated skin diseases31, revealed transition signatures from healthy to periodontal disease32, and offered specific signatures in the early and middle stages of human pregnancy with term birth outcome33.
Currently, only the serial quantitative assay of hCG and transvaginal ultrasound are the major diagnostic methods for EP34. Although these tools have enhanced the clinician’s ability to detect the disease, repeated evaluation is often required. Nevertheless, early diagnosis still remains a challenge, causing delay in disease management35. By invasive procedures, several biochemical markers have been studied and used for diagnosis by clinicians which include activin A, activin B, inhibin A, follistatin, A disintegrin and metalloprotease-12 (ADAM), pregnancy-associated plasma protein A (PAPP-A), pregnancy-specific B1-glycoprotein (SP1), interleukins 6 and 8 (IL-6 and IL 8), placental-like growth factor (PGF), vascular endothelial growth factor (VEGF), glycodelin (Glyc), etc.36,37,38,39,40,41,42. The ratio of EP to live births has been shown to increase with maternal age, and this ratio increased significantly from 11.0 to 13.7 ectopic pregnancies per 1000 live births between 2006 and 20136. Barnhart43 found that about half the percentage of people diagnosed with EP do not possess identifiable risk factors, including sexually transmitted infections, tubal damage, and pelvic inflammatory disease. Defects in ciliary movement and/or muscular contractions of fallopian tube, which facilitate transportation of embryo caused by infection or smoking, are the possible pathogenic mechanism for tubal ectopic pregnancy44. Reduction in adrenomedullin, a receptor protein, is a major factor that impairs embryo transport in fallopian tube and, thus, leads to tubal EP45,46. Many molecular approaches, including proteomics, have been explored for biomarker discovery for ectopic pregnancy. In a recent study, GSTO1 has been identified as a favorable protein biomarker from serum47. From the perspective of non-invasive approach, there has been no critical lead for salivary protein biomarker(s) for EP. Thus, we hypothesized that one or more salivary proteins would be potential non-invasive biomarker(s) to diagnose ectopic pregnancy. Therefore, this study attempted to explore the profile of salivary proteins during ectopic pregnancy by adopting gel-based protein mass spectrometry.
Materials and methods
Sample collection and processing
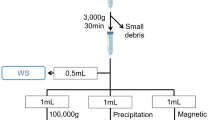
After obtaining proper informed consent from the volunteers, the saliva samples were collected as per the protocol approved by the Institutional Human Ethical Committee of Ravindra Nath Tagore (RNT) Medical College, Udaipur, India. All the procedures were carried out in accordance with the ethical guidelines and regulations. The unstimulated saliva samples were collected and assigned to four categories viz., (i) REP (ruptured ectopic pregnancy, n = 23); (ii) UREP (un-ruptured ectopic pregnancy, n = 17); (iii) PR (viable intra-uterine pregnancy, n = 15); and (iv) NPR (non-pregnant/control, n = 14). The mean age of the participants in the study was 27.47 ± 1.89 years for both ruptured and unruptured cases. In the case of pregnant and non-pregnant women, the mean age was 25.45 ± 2.96 years. Saliva collection was performed in the morning between 8 and 9 AM. Each volunteer was asked to abstain from food intake for 2 h before sampling and instructed for oral wash with sterile MilliQ water just before sampling. The saliva was collected by spitting method48 where the saliva was allowed to accumulate in the floor of the buccal cavity, and the volunteers spit out the saliva into sterile Falcon tubes every 60 s until the volume reached 2 mL. The collected samples were transported in a mini cooler box to the laboratory within 30 to 45 min after collection. An inhibitor cocktail was used to mitigate protease degradation during sample handling and transportation. The samples were then centrifuged at 4500×g for 15 min at 4 °C to remove the insoluble material and cells, if any, and then stored at − 80 °C until further use49.
Protein precipitation
To precipitate the salivary proteins, TCA-acetone precipitation method50 was employed. First, the saliva sample was mixed with 10% TCA and 10 mM DTT in acetone, and allowed to incubate for an hour at − 20 °C. Subsequently, the samples were subjected to centrifugation at 5000×g at 4 °C for 20 min. The resulting pellets were washed twice with ice-cold acetone and centrifuged each time at 5000×g at 4 °C for 20 min. Finally, the pellets were air-dried and re-suspended in 50 mM Tris buffer (pH 8.8). The protein concentration was determined using Bradford method51.
SDS-PAGE
To identify the salivary proteins, the samples were subjected to 12% SDS-PAGE analysis. A total of 30 μg of protein was taken from each individual sample and samples from each group were pooled separately. Subsequently, 30 μg of protein from the pooled salivary protein samples was mixed with sample buffer [50 mM Tris–HCl (pH 6.8), 2% SDS, 10% glycerol, 0.1% bromophenol blue, 100 mM β-mercaptoethanol] and heated at 60 °C for 1 min to ensure complete protein denaturation. The resultant mixture was loaded onto the gel, and electrophoresis was conducted at a constant current of 50 V. Following electrophoresis, the gels underwent a 2-min immersion in distilled water, followed by staining with 0.5% Coomassie Brilliant Blue (CBB) solution (composed of 40% methanol, 10% acetic acid, and 0.5% CBB R-250) at 37 °C for 2 h. Subsequently, a solution of 40% methanol and 10% acetic acid was used to de-stain the gels. Samples were pooled and quantified for group-wise REP, UREP, PR, and NPR, respectively, for LC–MS/MS analysis.
In-gel trypsin digestion
Trypsin digestion for separation of proteins in gels was conducted according to Shevchenko et al.52. The protein bands in the gel were treated with acetonitrile to remove the stain. Then, the gels were sliced and subjected to reduction using dithiothreitol (DTT) and ammonium bicarbonate. Next, the gel pieces were alkylated with iodoacetamide, washed and then incubated with trypsin, overnight, at 37 °C. After digestion, the peptides were extracted using formic acid and acetonitrile. The extracted fractions were dried and stored for further LC–MS/MS analysis at − 20 °C.
Peptide and protein identification by mass spectrometry
The peptides were desalted using C18 Pierce® Zip tips (Thermo Fisher Scientific, Waltham, MA, USA) and the digested peptide mixture was transferred to the vials in the LC autosampler. The peptides were ionized by nanospray capillary column (PepMap™ RSLC C18, Thermo Fisher Scientific, Waltham, MA, USA) and subjected to tandem mass spectrometry (MS/MS) on Q-Exactive HF (Thermo Fisher Scientific, Waltham, MA, USA). The threshold of false discovery rate was kept at 0.01. The MS/MS spectra were analyzed using Proteome Discoverer (Version 2.2) for protein identification. The MS/MS search was conducted using the SEQUEST search engine against the NCBI using Homo sapiens protein database. The search parameters included trypsin as a protease with one missed cleavage allowed. Carbamidomethyl cysteine was considered as a fixed modification, and oxidation of methionine was considered as a dynamic modification. The precursor ion mass error window was set at 10 ppm, and the fragment ion mass error window was set at 0.2 Da. To estimate the false discovery rate (FDR), peptide sequence analysis using a decoy database was enabled. High-confidence peptide identifications were obtained by setting a target FDR threshold of 1% at the peptide level.
Functional annotation
The identified proteins (UniProtKB Accession) were subjected to functional enrichment analysis using databases. Briefly, comprehensive information about the evolution of protein-coding gene families (particularly, protein phylogeny), their function, and the impact due to genetic variation were characterized using PANTHER (v16.0) classification system. Venn diagram was created by using web-based visualization tools viz., ProteoRE (http://proteore.org/). The identified proteins were curated by database of pathways and reactions in human biology for frequent cross-referencing of other resources [NCBI, Ensembl, UniProt, KEGG (gene and compound), ChEBI, PubMed, and GO] using Reactome (https://reactome.org/PathwayBrowser/#TOOL=AT). The proteome Reactome-annotated data describe the possible reactions if all annotated proteins and small molecules are present in a cell and active simultaneously53. A binomial test calculates the probability for each result. The p-values were corrected for the multiple testing (Benjamini–Hochberg procedure) that arise from evaluating the submitted list of identifiers against every pathway54.
Ethical statement
The study and the procedure followed in the sample collection were approved by the Institutional Human Ethics Committee (IHEC) (No. RNT/Stat/IEC/2019) of Ravindra Nath Tagore (RNT) Medical College, Udaipur.
Results
Total salivary proteome
The electrophoretically separated salivary proteins from various groups were subjected to mass spectrometry analysis (Fig. S1). A total of 326 proteins were identified across all groups of salivary samples. Among these 271, 166, 145 and 119 proteins were identified in REP, UREP, PR and NPR, respectively (Fig. 1). Further exploration of these 326 identified proteins, specifically 101 and 24 were found in REP (Table 1) and UREP (Table 2), respectively. In addition, 73 proteins were common to all four groups. Apart from REP and UREP, the proteins identified in the other groups are listed (Tables S1–S2).
Representation of the salivary proteins identified in ruptured ectopic pregnancy, un-ruptured ectopic pregnancy, normal pregnancy, and non-pregnancy (REP, UREP, PR, NPR). Venn diagram was drawn using web-based visualization tools viz., ProteoRE (A) (http://proteore.org/).
Functional annotation
The collected Uniprot GO terms by the identified proteins were subjected to functional annotation using PANTHER and Proteore databases. Proteins specific to REP were classified by their biological process, cellular components, and molecular function. Based on the biological process most of the proteins were related to cellular process (23.9%), whereas others were involved in metabolic processes (21.4%), biological regulation (11.9%), response to stimuli (11.4%), and immune system processes (8.5%), localization, signaling, interspecies interaction, multicellular organismal process, biological adhesion, developmental process, reproductive process, multi-organism process, and reproduction. Based on the cellular components, most of the salivary proteins got assigned to cellular anatomical entity (57.8%), intracellular (26.7%), and protein-containing complex (15.5%), respectively. According to the molecular function, the most prevalence was catalytic activity (46.7%), and binding (41.3%), followed by molecular function regulation, molecular transducer activity, translation regulator activity, and transporter activity, respectively (Fig. 2).
Pathway enrichment analysis
Reactome analysis was performed using the resources from NCBI, Ensembl, UniProt, KEGG, ChEBI, PubMed, and GO, which showed 81 out of the 101 identified proteins to be found in Reactome knowledge, where 485 pathways were hit at least once. About 25 significant pathways were sorted with p-value (Table S3). Further analysis revealed that two proteins, plasminogen activator inhibitor 2 (SERPINB2) and plasma protease C1 inhibitor (SERPING1) are specifically involved in coagulation and complement cascade pathways, respectively (Fig. 3).
Protein–protein interaction
Protein–protein interaction network was constructed by retrieving the String, and the results showed that 87 proteins were in connection with other proteins, which lead to 204 paired relationships in medium interaction score, which indicates moderate confidence in the detected protein–protein interactions. Additionally, the proteins related to 102 relationships in high interaction score (Fig. 4), which are also part of the medium scoring pairs. The scores were calculated based on the data collected from experimental computational prediction with a confidence value. Among the proteins, QSOX1, CHI3L1, CHIT1, CTSZ, MMP8, PGM1, GSTO1, FN1 and LTA4H showed strong interactions among themselves and also with other proteins in the network.
String representative network of the identified REP-specific salivary proteins. (A) The minimum required interaction score is set as 0.4. A total of 87 genes were connected with 204 paired relationships annotated. (B) The minimum required interaction score was set as 0.9. A total of 87 genes were related to 102 paired relationships. The PPI enrichment p-value: < 1.0e−16. No relationship nodes were removed from the network.
Discussion
In the present study, numerous immunological and non-immunological proteins have been detected in the saliva of women representing different conditions viz., REP (ruptured ectopic pregnancy; Group 1), UREP (un-ruptured ectopic pregnancy; Group 2), PR (pregnancy; Group 3) and NPR (non-pregnancy, Group 4). Specifically, REP saliva contains many defensive proteins which throw new light on the understanding of the defense mechanisms mediated by saliva during ectopic pregnancy. Also, these proteins have been reported to possess antibacterial activity55.
Fibronectin (FN1) is a protein found in the extracellular matrix (ECM) that regulates cell adhesion, spreading, migration, proliferation, and apoptosis56. In pregnant women, fetal fibronectin (fFN) plays an important role in the pathogenesis of preterm labor and premature rupture of fetal membranes57,58. Possibly, one of the salivary proteins in this study is fFN that has earlier been identified in the saliva of exclusively EPR cases. The expression of vitamin D-binding protein is lower in the placenta59 and has been found in serum60, leading to spontaneous miscarriages and preterm deliveries, respectively. Interestingly, in the present study vitamin D shows up in the salivary samples at REP. Another protein, known as apolipoprotein B-100 (ApoB), the level of which is higher in preeclampsia (PE), fetal growth restriction (FGR), and PE + FGR than the normal pregnancy61. Fibronectin, vitamin D-binding protein, and ApoB are found in saliva specifically during the REP which reflects that saliva has the potential to showcase the progress in ectopic pregnancy condition. The protein ezrin is identified in REP condition, which is an interesting observation since Ezrin and its activated form have been observed in endometriotic lesions, evidencing that these proteins could play some role in the migration and attachment of endometriotic lesions62.
Human saliva contains several Cystatin family proteins, including Cystatin-A, Cystatin-B, Cystatin-C, Cystatin-D, Cystatin-S, Cystatin-SA and Cystatin-SN63. Particularly appearance of Cystatin-S, a cysteine protease inhibitor, is prominent during the ovulatory phase28. However, in the present study, Cystatin-A appeared during REP condition. Several proteins, such as Alpha-1-acid glycoprotein 2, Complement C4-B, Immunoglobulin heavy variable 1–2, Immunoglobulin heavy variable 1–3, Immunoglobulin heavy variable 1–46, Prostatic acid phosphatase, and Vitamin D-binding protein have been identified in the human follicular fluid64. Interestingly, these proteins are present in the saliva of women during REP condition and completely absent in PR and NPR saliva.
Previous research has provided valuable insights into the immune mechanisms involved in reproductive health, shedding light on the role of the immune system in the context of fallopian tubes65. Additionally, Wicherek et al.66,67 reported increased levels of CD56+ and CD3+ cells, as well as heightened CD69 staining, in ruptured ectopic pregnancies. These findings suggest immune cell involvement in the process of tubal rupture, which is crucial information for understanding the pathogenesis of ectopic pregnancies. Furthermore, Visser et al.68 discussed the historical use of anti-Rh(D) immunoglobulin in Rh-negative women as a preventive measure against sensitization. This emphasizes the importance of immune-related interventions in reproductive health management. However, it is noteworthy that the current evidence on immune cell subtypes in tubal pathologies remains inconsistent. Only limited conclusions can be drawn at this stage, emphasizing the necessity for further research to explore and clarify the role of specific immune cell subtypes in tubal pathologies.
Notably, Complement C4-B is reportedly upregulated in the follicular fluid of patients with recurrent spontaneous abortion69, and its potential presence in saliva may have implications for ectopic pregnancy (EP) research. Additionally, proteins such as Plasminogen Activator Inhibitor 2 and 1, expressed in human saliva, may also be relevant to EP. The low expression rate of plasminogen activator inhibitor type-2 (PAI-2) leads to an increased risk of growth restriction in developing intrauterine layers70.
The pathways enrichment encoding genes-associated biological pathways are concerned with immunity, inflammation, homeostasis, and development. Importantly, plasminogen activator inhibitor 2 (SERPINB2) and plasma protease C1 inhibitor (SERPING1) are involved in coagulation and complement cascade which may throw open use as a putative salivary biomarker protein for diagnosis of REP condition. It has been emphasized that preeclampsia and eclampsia are two pregnancy complications that are basic to occurrence of REP is the presence of placental tissue in the maternal body and it is postulated that poor placentation results from inappropriate uterine spiral artery invasion65. The low level of SERPINB2 is associated with placental insufficiency, and the high level of tissue plasminogen activator is connected to endothelial dysfunction in patients with severe preeclampsia71. Low-level plasma protease C1 inhibitor has been used as a biomarker for preeclampsia72. The present study also finds that salivary proteins could be potential biomarkers of ectopic pregnancy, a subject worthy of further investigation.
Glutathione S-transferase omega-1 (GSTO1) is an enzyme that plays an important role in detoxification of environmental pollution and in xenobiotic metabolism. Also, the level of GSTO1 is related to fetal intrauterine growth restriction. Glutathione S-transferase omega-1 has been identified as a potential biomarker in a data-independent acquisition (DIA) proteomics study to evaluate ectopic pregnancy (EP) using serum samples. However, the study found that GSTO1 levels were significantly different between the intrauterine pregnancy (IP) and EP groups47. Functional annotation revealed that the salivary proteins of interest as above are majorly associated with binding property and regulatory function in REP and UREP conditions. Regulatory proteins activate or repress transcription. Activators enhance RNA polymerase interaction with the promoter, while repressors impede progress. These proteins are crucial for proper organ development by turning on specific genes at the right time. Protein–protein interaction analysis shows a crosstalk among QSOX1, CHI3L1, CHIT1, CTSZ, MMP8, PGM1, GSTO1, FN1, and LTA4H. These interactions have a significant role during miscarriage. Based on the reports these nine proteins are predominantly associated with ectopic pregnancy, which is recaptured in the present study.
Considering other regular body fluids, saliva has potential as a source of biomarker proteins. Since there are no remarkable reports on proteomic studies to show the variation of proteins present in saliva with respect to ectopic pregnancy condition by adopting proteins mass spectrometry technology, to the best of our knowledge, this is the first mass-spectrometry-based proteomic study of saliva to explore the salivary proteins focusing on ectopic pregnancy condition. Among the identified 271 proteins, the 101 REP-specific proteins offer the potential to spot out the promising protein biomarkers. Further studies on the immunoglobulin, fibronectin, vitamin D-binding protein, plasminogen activator inhibitor 2, ApoB, and Cystatin-A can potentially lead to selection of putative candidate salivary biomarkers of REP condition in women. Further, the functional annotation of salivary proteins strongly indicates that the proteins of interest are mostly extracellular proteins that participate in regulatory functions during REP and UREP conditions. Ectopic pregnancy, being a tissue-specific risk factor, identification of the panel of protein markers in the saliva may help in development of non-invasive/cost-affordable diagnostic tool for the EP. Moreover, our preliminary data suggests that salivary proteins, including Complement C4-B, Plasminogen Activator Inhibitor 2 and 1, and Glutathione S-transferase omega-1, hold promise as potential biomarkers for ectopic pregnancy. These proteins have been implicated in other issues in reproduction, and show potential relevance to ectopic pregnancy. A more elaborate and focused study, currently in progress, is expected to throw light on the specificity and diagnostic utility of the specific salivary protein(s) associated with ectopic pregnancy.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036633.
References
Heinberg, E. M., McCoy, T. W. & Pasic, R. The perforated intrauterine device: Endoscopic retrieval. JSLS 12, 97–100 (2008).
Ward, Z. J., Atun, R., King, G., Sequeira Dmello, B. & Goldie, S. J. Simulation-based estimates and projections of global, regional and country-level maternal mortality by cause, 1990–2050. Nat. Med. 29, 1253–1261. https://doi.org/10.1038/s41591-023-02310-x (2023).
Hendriks, E., Rosenberg, R. & Prine, L. Ectopic pregnancy: Diagnosis and management. Am. Fam. Physician 101, 599–606 (2020).
Meh, C. et al. Trends in maternal mortality in India over two decades in nationally representative surveys. BJOG Int. J. Obstet. Gynaecol. 129, 550–561. https://doi.org/10.1111/1471-0528.16888 (2021).
Gertie Makhado, T., Makhado, L., Lizzy Netshikeweta, M. & Rose Azwidihwi, T. in Midwifery: New Perspectives and Challenges [Working Title] (2023).
Mann, L. M., Kreisel, K., Llata, E., Hong, J. & Torrone, E. A. Trends in ectopic pregnancy diagnoses in United States emergency departments, 2006–2013. Matern. Child. Health J. 24, 213–221. https://doi.org/10.1007/s10995-019-02842-0 (2020).
Tahmina, S., Daniel, M. & Solomon, P. Clinical analysis of ectopic pregnancies in a tertiary care centre in Southern India: A six-year retrospective study. J. Clin. Diagn. Res. 10, 13–16. https://doi.org/10.7860/JCDR/2016/21925.8718 (2016).
Pal, A., Gupta, K. B. & Sarin, R. A study of ectopic pregnancy and high risk factors in Himachal Pradesh. J. Indian Med. Assoc. 94, 172–173 (1996).
Dvash, S. et al. Increase rate of ruptured tubal ectopic pregnancy during the COVID-19 pandemic. Eur. J. Obstet. Gynecol. Reprod. Biol. 259, 95–99. https://doi.org/10.1016/j.ejogrb.2021.01.054 (2021).
Humphrey, S. P. & Williamson, R. T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 85, 162–169. https://doi.org/10.1067/mpr.2001.113778 (2001).
Amado, F., Lobo, M. J., Domingues, P., Duarte, J. A. & Vitorino, R. Salivary peptidomics. Expert Rev. Proteomics 7, 709–721. https://doi.org/10.1586/epr.10.48 (2010).
Amado, F. M., Vitorino, R. M., Domingues, P. M., Lobo, M. J. & Duarte, J. A. Analysis of the human saliva proteome. Expert Rev. Proteomics 2, 521–539. https://doi.org/10.1586/14789450.2.4.521 (2005).
Meyer-Lueckel, H., Hopfenmuller, W., von Klinggraff, D. & Kielbassa, A. M. Microradiographic study on the effects of mucin-based solutions used as saliva substitutes on demineralised bovine enamel in vitro. Arch. Oral Biol. 51, 541–547. https://doi.org/10.1016/j.archoralbio.2006.01.006 (2006).
Niswander, J. D., Shreffler, D. C. & Neel, J. V. Genetic studies of quantitative variation in a component of human saliva. Ann. Hum. Genet. 27, 319–328. https://doi.org/10.1111/j.1469-1809.1963.tb01529.x (1964).
Huang, A. Y., Castle, A. M., Hinton, B. T. & Castle, J. D. Resting (Basal) secretion of proteins is provided by the minor regulated and constitutive-like pathways and not granule exocytosis in parotid acinar cells. J. Biol. Chem. 276, 22296–22306. https://doi.org/10.1074/jbc.M100211200 (2001).
Zambonin, C. & Aresta, A. MALDI-TOF/MS analysis of non-invasive human urine and saliva samples for the identification of new cancer biomarkers. Molecules 27, 1972. https://doi.org/10.3390/molecules27061925 (2022).
Liu, J. et al. Saliva diagnostics: Emerging techniques and biomarkers for salivaomics in cancer detection. Expert Rev. Mol. Diagn. 22, 1077–1097. https://doi.org/10.1080/14737159.2022.2167556 (2022).
Nam, Y., Kim, Y. Y., Chang, J. Y. & Kho, H. S. Salivary biomarkers of inflammation and oxidative stress in healthy adults. Arch. Oral Biol. 97, 215–222. https://doi.org/10.1016/j.archoralbio.2018.10.026 (2019).
Ryu, O. H., Atkinson, J. C., Hoehn, G. T., Illei, G. G. & Hart, T. C. Identification of parotid salivary biomarkers in Sjogren’s syndrome by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry and two-dimensional difference gel electrophoresis. Rheumatology 45, 1077–1086. https://doi.org/10.1093/rheumatology/kei212 (2006).
Xiao, H. et al. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell Proteomics 11, 111012112. https://doi.org/10.1074/mcp.M111.012112 (2012).
Hu, S. et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 56, 3588–3600. https://doi.org/10.1002/art.22954 (2007).
Xiao, H. & Wong, D. T. Proteomics and its applications for biomarker discovery in human saliva. Bioinformation 5, 294–296. https://doi.org/10.6026/97320630005294 (2011).
Mahajan, P. G. et al. Comparison of salivary total protein and electrolyte profile in HIV patients with and without antiretroviral therapy. Dis. Mon. 67, 101165. https://doi.org/10.1016/j.disamonth.2021.101165 (2021).
Zhang, N. et al. Quantitative analysis of differentially expressed saliva proteins in human immunodeficiency virus type 1 (HIV-1) infected individuals. Anal. Chim. Acta 774, 61–66. https://doi.org/10.1016/j.aca.2013.02.038 (2013).
Gibbins, H. L., Proctor, G. B., Yakubov, G. E., Wilson, S. & Carpenter, G. H. Concentration of salivary protective proteins within the bound oral mucosal pellicle. Oral Dis. 20, 707–713. https://doi.org/10.1111/odi.12194 (2014).
Bencharit, S. et al. Salivary proteins associated with hyperglycemia in diabetes: A proteomic analysis. Mol. Biosyst. 9, 2785–2797. https://doi.org/10.1039/c3mb70196d (2013).
Saibaba, G. & Archunan, G. Does salivary protein(s) act an ovulation indicator for women? A hypothesis. Med. Hypotheses 84, 104–106. https://doi.org/10.1016/j.mehy.2014.12.009 (2015).
Saibaba, G. et al. Salivary proteome profile of women during fertile phase of menstrual cycle as characterized by mass spectrometry. Gynecol. Minim. Invasive Ther. 10, 226–234. https://doi.org/10.4103/GMIT.GMIT_78_20 (2021).
Katsani, K. R. & Sakellari, D. Saliva proteomics updates in biomedicine. J. Biol. Res. 26, 17. https://doi.org/10.1186/s40709-019-0109-7 (2019).
Pappa, E. et al. Saliva proteomics analysis offers insights on type 1 diabetes pathology in a pediatric population. Front. Physiol. 9, 444. https://doi.org/10.3389/fphys.2018.00444 (2018).
Campanati, A. et al. Saliva proteomics as fluid signature of inflammatory and immune-mediated skin diseases. Int. J. Mol. Sci. 22, 7018. https://doi.org/10.3390/ijms22137018 (2021).
Bostanci, N. et al. Targeted proteomics guided by label-free quantitative proteome analysis in saliva reveal transition signatures from health to periodontal disease. Mol. Cell Proteomics 17, 1392–1409. https://doi.org/10.1074/mcp.RA118.000718 (2018).
Dey, A. K. et al. Salivary proteome signatures in the early and middle stages of human pregnancy with term birth outcome. Sci. Rep. 10, 8022. https://doi.org/10.1038/s41598-020-64483-6 (2020).
Alkatout, I. et al. Clinical diagnosis and treatment of ectopic pregnancy. Obstet. Gynecol. Surv. 68, 571–581. https://doi.org/10.1097/OGX.0b013e31829cdbeb (2013).
Farquhar, C. M. Ectopic pregnancy. Lancet 366, 583–591. https://doi.org/10.1016/S0140-6736(05)67103-6 (2005).
Florio, P. et al. Serum activin A levels are lower in tubal than intrauterine spontaneously conceived pregnancies. Gynecol. Endocrinol. 27, 391–395. https://doi.org/10.3109/09513590.2010.495430 (2011).
Rausch, M. E. et al. Development of a multiple marker test for ectopic pregnancy. Obstet. Gynecol. 117, 573–582. https://doi.org/10.1097/AOG.0b013e31820b3c61 (2011).
Warrick, J. et al. Serum activin A does not predict ectopic pregnancy as a single measurement test, alone or as part of a multi-marker panel including progesterone and hCG. Clin. Chim. Acta 413, 707–711. https://doi.org/10.1016/j.cca.2011.12.018 (2012).
Daponte, A. et al. Activin A and follistatin as biomarkers for ectopic pregnancy and missed abortion. Dis. Markers 35, 497–503. https://doi.org/10.1155/2013/969473 (2013).
Senapati, S. et al. Predicting first trimester pregnancy outcome: Derivation of a multiple marker test. Fertil. Steril. 106, 1725–1732. https://doi.org/10.1016/j.fertnstert.2016.08.044 (2016).
Dhiman, P. et al. Serum activin B concentration as predictive biomarker for ectopic pregnancy. Clin. Biochem. 49, 609–612. https://doi.org/10.1016/j.clinbiochem.2015.11.024 (2016).
Rajendiran, S. et al. Diagnostic significance of IL-6 and IL-8 in tubal ectopic pregnancy. J. Obstet. Gynaecol. 36, 909–911. https://doi.org/10.1080/01443615.2016.1174821 (2016).
Barnhart, K. T. Clinical practice: Ectopic pregnancy. N. Engl. J. Med. 361, 379–387. https://doi.org/10.1056/NEJMcp0810384 (2009).
Ankum, W. M., Mol, B. W., Van der Veen, F. & Bossuyt, P. M. Risk factors for ectopic pregnancy: A meta-analysis. Fertil. Steril. 65, 1093–1099 (1996).
Wang, X. et al. Risk factors and clinical characteristics of recurrent ectopic pregnancy: A case-control study. J. Obstet. Gynaecol. Res. 46, 1098–1103. https://doi.org/10.1111/jog.14253 (2020).
Li, C. et al. Risk factors for ectopic pregnancy: A multi-center case-control study. BMC Pregnancy Childbirth 15, 187. https://doi.org/10.1186/s12884-015-0613-1 (2015).
Ma, D. et al. Identification of noninvasive diagnostic biomarkers for ectopic pregnancy using data-independent acquisition (DIA)proteomics: A pilot study. Sci. Rep. 12, 19992. https://doi.org/10.1038/s41598-022-23374-8 (2022).
Navazesh, M. Methods for collecting saliva. Ann. N. Y. Acad. Sci. 694, 72–77. https://doi.org/10.1111/j.1749-6632.1993.tb18343.x (1993).
Jessie, K., Pang, W. W., Haji, Z., Rahim, A. & Hashim, O. H. Proteomic analysis of whole human saliva detects enhanced expression of interleukin-1 receptor antagonist, thioredoxin and lipocalin-1 in cigarette smokers compared to non-smokers. Int. J. Mol. Sci. 11, 4488–4505. https://doi.org/10.3390/ijms11114488 (2010).
Koontz, L. Laboratory Methods in Enzymology: Protein Part C Methods in Enzymology, 3–10 (2014).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. https://doi.org/10.1006/abio.1976.9999 (1976).
Shevchenko, A. et al. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24, 893–896. https://doi.org/10.1042/bst0240893 (1996).
Jassal, B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503. https://doi.org/10.1093/nar/gkz1031 (2020).
Jalili, V. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48, W395–W402. https://doi.org/10.1093/nar/gkaa434 (2020).
de Almeida, P. V., Gregio, A. M., Machado, M. A., de Lima, A. A. & Azevedo, L. R. Saliva composition and functions: a comprehensive review. J. Contemp. Dent. Pract. 9, 72–80 (2008).
Chen, Y. et al. Overdosage of balanced protein complexes reduces proliferation rate in aneuploid cells. Cell Syst. 9, 129–142. https://doi.org/10.1016/j.cels.2019.06.007 (2019).
Ruma, M. S., Bittner, K. C. & Soh, C. B. Current perspectives on the use of fetal fibronectin testing in preterm labor diagnosis and management. Am. J. Manag. Care 23, S356–S362 (2017).
Blackwell, S. C. et al. Utilization of fetal fibronectin testing and pregnancy outcomes among women with symptoms of preterm labor. Clin. Outcomes Res. 9, 585–594. https://doi.org/10.2147/CEOR.S141061 (2017).
Hou, H. et al. Altered decidual and placental catabolism of vitamin D may contribute to the aetiology of spontaneous miscarriage. Placenta 92, 1–8. https://doi.org/10.1016/j.placenta.2020.01.013 (2020).
D’Silva, A. M., Hyett, J. A. & Coorssen, J. R. First trimester protein biomarkers for risk of spontaneous preterm birth: Identifying a critical need for more rigorous approaches to biomarker identification and validation. Fetal Diagn. Ther. 47, 497–506. https://doi.org/10.1159/000504975 (2020).
Alahakoon, T. I., Medbury, H. J., Williams, H. & Lee, V. W. Lipid profiling in maternal and fetal circulations in preeclampsia and fetal growth restriction: A prospective case control observational study. BMC Pregnancy Childbirth 20, 61. https://doi.org/10.1186/s12884-020-2753-1 (2020).
Peloggia, A., Andres, M. P. & Abrao, M. S. Expression of ezrin protein and phosphorylated ezrin in pelvic endometriotic lesions. Clinics 77, 100074. https://doi.org/10.1016/j.clinsp.2022.100074 (2022).
Saibaba, G. et al. Proteomic analysis of human saliva: An approach to find the marker protein for ovulation. Reprod. Biol. 16, 287–294. https://doi.org/10.1016/j.repbio.2016.10.005 (2016).
Shen, X. et al. Proteomic analysis of human follicular fluid associated with successful in vitro fertilization. Reprod. Biol. Endocrinol. 15, 58. https://doi.org/10.1186/s12958-017-0277-y (2017).
Rigby, C. H. et al. The immune cell profile of human fallopian tubes in health and benign pathology: A systematic review. J. Reprod. Immunol. 152, 103646. https://doi.org/10.1016/j.jri.2022.103646 (2022).
Wicherek, L., Galazka, K. & Lazar, A. Analysis of metallothionein, RCAS1 immunoreactivity regarding immune cell concentration in the endometrium and tubal mucosa in ectopic pregnancy during the course of tubal rupture. Gynecol. Obstet. Invest. 65, 52–61. https://doi.org/10.1159/000107649 (2008).
Wicherek, L. et al. Metallothionein expression and infiltration of cytotoxic lymphocytes in uterine and tubal implantation sites. J. Reprod. Immunol. 70, 119–131. https://doi.org/10.1016/j.jri.2005.12.003 (2006).
Visser, G. H. A. et al. FIGO/ICM guidelines for preventing rhesus disease: A call to action. Int. J. Gynaecol. Obstet. 152, 144–147. https://doi.org/10.1002/ijgo.13459 (2021).
Sun, X. et al. Decreased histidine-rich glycoprotein and increased complement C4-B protein levels in follicular fluid predict the IVF outcomes of recurrent spontaneous abortion. Clin. Proteomics 19, 47. https://doi.org/10.1186/s12014-022-09383-9 (2022).
Coolman, M. et al. Angiogenic and fibrinolytic factors in blood during the first half of pregnancy and adverse pregnancy outcomes. Obstet. Gynecol. 119, 1190–1200. https://doi.org/10.1097/AOG.0b013e318256187f (2012).
Tanjung, M. T., Siddik, H. D., Hariman, H. & Koh, S. C. Coagulation and fibrinolysis in preeclampsia and neonates. Clin. Appl. Thromb. Hemost. 11, 467–473. https://doi.org/10.1177/107602960501100415 (2005).
Blankley, R. T. et al. A proof-of-principle gel-free proteomics strategy for the identification of predictive biomarkers for the onset of pre-eclampsia. BJOG 116, 1473–1480. https://doi.org/10.1111/j.1471-0528.2009.02283.x (2009).
Acknowledgements
G.A. acknowledges the University Grants Commission, New Delhi, India, for the award of UGC-BSR Faculty Fellow (No. F.18-1/2011(BSR) dt. 4 January 2017). The authors thank the volunteers from Ravindra Nath Tagore (RNT) Medical College, Udaipur, India, for effective participation in the study.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.P.A., S.S. and G.A.; Methodology & Formal Analysis: M.S. and A.P.A.; Writing-Original Draft: M.S., and S.M.; Data analysis, Writing, Review & Editing: A.P.A., S.S., M.S., S.M., P.M., M.A.A., and G.A.; Supervision: S.S. and G.A.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Priya Aarthy, A., Sen, S., Srinivasan, M. et al. Ectopic pregnancy: search for biomarker in salivary proteome. Sci Rep 13, 16828 (2023). https://doi.org/10.1038/s41598-023-43791-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-023-43791-7