Abstract
Accurate diagnosis of white blood cells from cytopathological images is a crucial step in evaluating leukaemia. In recent years, image classification methods based on fully convolutional networks have drawn extensive attention and achieved competitive performance in medical image classification. In this paper, we propose a white blood cell classification network called ResNeXt-CC for cytopathological images. First, we transform cytopathological images from the RGB color space to the HSV color space so as to precisely extract the texture features, color changes and other details of white blood cells. Second, since cell classification primarily relies on distinguishing local characteristics, we design a cross-layer deep-feature fusion module to enhance our ability to extract discriminative information. Third, the efficient attention mechanism based on the ECANet module is used to promote the feature extraction capability of cell details. Finally, we combine the modified softmax loss function and the central loss function to train the network, thereby effectively addressing the problem of class imbalance and improving the network performance. The experimental results on the C-NMC 2019 dataset show that our proposed method manifests obvious advantages over the existing classification methods, including ResNet-50, Inception-V3, Densenet121, VGG16, Cross ViT, Token-to-Token ViT, Deep ViT, and simple ViT about 5.5–20.43% accuracy, 3.6–23.56% F1-score, 3.5–25.71% AUROC and 8.1–36.98% specificity, respectively.
Similar content being viewed by others
Introduction
In recent years, an increase in the incidence of cancer in the world has risen sharply, and the number of patients is growing year by year. The amount of cancer patients worldwide has rocketed from 17.2 million in 2016 to 19.3 million in 2020. It is estimated that the amount will rise to 20.2 million in 20221. Of the cancers, leukaemia is the most common blood cancer and is thought to contribute to about 60,000 deaths every year2.
Early diagnosis of leukaemia can remarkably improve the survival rate. In medicine, the early diagnosis of leukaemia is usually made by clinicians observing the morphology and structure of bone marrow and blood cells, such as microscopic examination of bone marrow aspiration and blood smears3. The microscopic examination of blood smears usually requires clinical doctors to observe and measure manually under a microscope, which is tedious and time-consuming, and the diagnostic accuracy depends upon prior knowledge, contextual information and operator’s experience, and so on. Thus, accurate, automated and repeatable methods for white blood cell classification are urgently needed in clinical practice.
Accurate white blood cell classification from cytopathological images depends upon the effective extraction of characteristics of white blood cells. However, this is difficult for several reasons. First, different cells vary greatly in shape, size, edge and position. Second, different staining methods and colorants can give rise to color differences among similar cells in different images. Finally, digital acquisition from different instruments for the same blood smear will also produce large differences in color. Nowadays, a multitude of cell classification methods have been proposed, including traditional methods4,5,6,7,8,9, machine learning methods10,11,12 and deep learning methods13,14,15,16,17,18,19,20,21,22,23. However, the traditional methods and the machine learning methods require hand-crafted features, the extraction of which is cumbersome and time-consuming, and the results are usually unsatisfactory.
Recently, with the rapid advancement of artificial intelligence, deep learning technology has swiftly permeated the medical field. Convolutional neural networks (CNNs) possess stable and reliable modelling capabilities. They can automatically extract image features and integrate them to generate higher-level semantic features based on global information, thereby enhancing the efficiency of image processing.
In 2017, Hung et al.13 used the Faster R-CNN network for the first time on malaria-infected blood smear images to identify infected cells and distinguish different stages of infection according to different characteristics of the cells. In the same year, Duggal et al.14 carried out research on normal and malignant cell classification of white blood cells in blood smears. They transformed microscopic images from RGB color space to optical density space. According to the principle of cell staining, the optical density space reveals a significant number of stained cells, which can provide detailed the characteristics of the cells. In 2019, Shahin et al.15 presented a new identification system for five types of white blood cells based on CNN, including transfer learning and fine-tuning of existing deep networks. They also proposed an end-to-end WBCsNet. In 2020, Huang et al.16 proposed an attention-aware residual network based manifold learning model (ARML) to classify white blood cells. Their aim is to enhance the network’s learning ability for classifying six types of white blood cells. In 2021, Shi et al.17 proposed to use Graph Convolutional Network (GCN) for cervical cell classification. First, features of all cervical cell images were extracted and clustered by CNN, and then a graph structure was used to further capture the underlying correlations among clusters. Second, GCN was applied to propagate the node dependency, thus producing the relation-aware feature representation. Finally, GCN features were added to strengthen the discriminative ability of CNN for features. In 2022, Ha et al.18 proposed a semi-supervised white blood cell classification method, which contained an SSTS module and a FGIA mechanism. In 2022, Sharma et al.19 proposed a deep learning method, which ultilizes the DenseNet121 to classify the white blood cells. In 2023, Zheng et al.20 proposed a two-stage method, which treated WBC detection as a small salient object detection task. In the saliency detection stage, Itti’s visual attention model was used to locate the ROIs. In the WBC detection stage, a more precise WBC classification was obtained by performing the modified CenterNet model on ROI sub-images. In the same year, Tummala et al.21 proposed a Siamese twin network (STN) model based on contrastive learning for the classification of normal peripheral blood cells, which was trained on relatively few images and used a EfficientNet-B3 as the backbone network. In 2024, Dhalla et al.22 proposed a model that simulated the real human brain, which consisted of many convolutional layers, each having its own set of additional skip connections. Useage of vector made them rotationally invariant. Eventually, the network was simplified by 20%, facilitating its deployment in resource-constrained environments. Gao et al.23 proposed a generative adversarial network (GAN) assisted framework, which can enhance images and boost the performance of peripheral leukocyte recognition and classification. However, many of the exiting methods are not sufficiently tailored to the specific characteristics of white blood cell images. The current network's ability to extract features is insufficient for distinguishing between normal and tumor cells and needs to be further improved.
Although deep learning based cell classification methods have made far more achievements than other methods, there are still some challenges in cell classification research as follows:
-
1.
Current classification models for white blood cell discrimination are usually designed to implement cell classification directly. However, as there are various types of cells in blood smear images, white blood cell classification on blood smear images is easily disturbed by the background, resulting in unsatisfactory classification results.
-
2.
The morphological characteristics of normal and tumor cells are very similar, and only local characteristics are slightly different. This is a fine-grained classification task, which is more difficult than the common classification tasks.
To address the above problems, motivated by the ResNeXt classification network24, we propose a ResNeXt-Cell classification (ResNeXt-CC) network for white blood cell classification from cytopathological images. Our study makes the following contributions:
-
1.
Our study reveals that a suitable color space model can highlight the dynamic contrast and texture details of white blood cells. That is to say, the transformation of the color space can suppress or enhance the expression of some characteristics of images, thus promoting the classification performance.
-
2.
Given that convolutions on different levels contain different discriminative information, and for the fine-grained classification task, local features play an important role in distinguishing different classes.We propose a cross-layer deep-feature fusion module (CFFM), which aims to fuse the discriminative feature information from different layers to improve the network performance.
-
3.
To avoid the negative effects of feature compression and enhance cross-channel information extraction. ECANet is introduced into our network to enhance discriminative information extraction. ECANet 0 is introduced into our network, aiming at enhancing the extraction ability of discriminative information, thereby improving the classification results.
-
4.
Since the cells morphology is highly similar, only the local characteristics differ, and there is a class imbalance between normal and tumor cells, we improve the softmax loss function and combine it with the center loss function to maximize the inter-class separation and enhance the intra-class compactness.
Methods
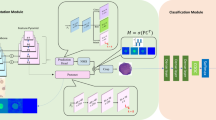
We propose a ResNeXt-CC network, as shown in Fig. 1. First, the efficient attention mechanism of ECANet module is used to strengthen feature extraction ability of cell details; second, a CFFM is proposed to enhance the discriminative information extraction; finally, we improve the softmax loss function and introduce the center loss function to train the network, which effectively addresses the problem of class imbalance and improves the discriminative information extraction.
ResNeXt classification model
Due to the small size of cytopathological image datasets, only increasing the network depth makes the network too complex, thus leading to the network over-fitting. The ResNeXt architecture can better extract features and achieve higher classification accuracy with fewer network parameters by combining ResNet-5026 and Inception27.
In this paper, the ResNeXt-50 realizes feature extraction through multiple ResNeXt modules and down-sampling layers, and then classifies images through the fully connected layer. Considering that it is relatively simple to extract features in the pooling layer, which may miss some effective information, and using a convolution method to down-sample can retain more information. Therefore, the convolution layer is used for down-sampling in our classification method instead of the pooling layer. The detailed architecture of the ResNeXt-50 model in this paper is shown in Table 1, where C represents the number of groups, x of Conv5_x represents the x-th ResNeXt module.
Data enhancement for cytopathological images
In the field of medical imaging, due to the diversity of detection methods and equipment, different image features differ greatly. Therefore, image enhancement methods are designed according to the characteristics of different medical images. Particularly, color space transformation is an image enhancement method, that is, some images with low contrast and unclear meaning are transformed into other color spaces. This transformation can suppress or enhance the expression of some characteristics of images. In this paper, we use color space transformation to preprocess cytopathological images, with the aim of making it easier for the network to extract important information, to improve the network performance.
The RGB color model is widely used in medical imaging. The superposition of the red, green and blue light components in different proportions generates different colors. Moreover, image brightness is also closely related to these three components. In contrast, the HSV color model comprises three distinct color bands: hue (H), saturation (S), and lightness (V), where H represents color information, and its value ranges from 0° to 360°. S value ranges from 0 to 1: the higher the value, the darker and brighter the color. V value ranges from 0 to 1: the higher the value, the higher the brightness. In our experiment, prior to model training, the values within each channel of the HSV image representation were normalized into the range of [0, 1].
The HSV color space has more advantages over the RGB color space in terms of expressing hue, brightness and so on. In cytopathological images, different components of stained cells present different colors. Compared with the RGB color space, the HSV color space can better represent the hue, saturation, brightness and so on in cytopathological images. In this paper, the RGB color space is transformed into the HSV color space, which is more conducive to extracting cell details.
The image comparison before and after the transformation is shown in Fig. 2. We can see that the dynamic contrast and texture details of the latter are more prominent. Subsequent experiments show that the classification accuracy of white blood cells is significantly improved after the cytopathological images are transformed from the RGB to the HSV color spaces.
ResNeXt-CC cell classification method
Cross-layer deep-feature fusion module
Although normal white blood cells and tumor white blood cells have high morphological similarity, their local characteristics are slightly different. Therefore, it is necessary to strengthen the network to identify the difference of local characteristics between classes to improve classification accuracy.
It is worth noting that different semantic information can be extracted from convolutional layers with different receptive fields, so there is a problem of information loss in the down-sampling process of CNN. Compared with common classification tasks, for the fine-grained classification tasks such as cell classification, using only one convolutional layer at the same network depth to obtain the characteristic information of cells often loses too much useful information. In recent years, many researchers have explored multi-level feature fusion. In Bilinear CNN28, two pooling operations were designed to fuse features from two branches to better obtain the spatial relationship between regions and improve classification accuracy. In addition, many feature fusion strategies have emerged in semantic segmentation and target detection fields. The U-net29 fused the feature information from different convolutional layers through the code-decode structure to achieve better segmentation results. The feature pyramid network30 used multi-scale feature fusion to further improve the detection accuracy. To sum up, a multitude of methods based on feature fusion have achieved good results in various fields of deep learning.
In this section, to explore the different discriminative feature information contained in different convolutional layers, the Grad-CAM31 algorithm is used to visualize the last layer of the Conv5_1, Conv5_2 and Conv5_3 modules from the backbone network, and the visualization results are shown in Fig. 3. The redder areas in the heatmaps indicate that the network pays more attention to them. From Fig. 3, the different convolutional layers from the classification network pay attention to different areas of cells and contain different discriminative feature information.
Based on the above analyses, to better obtain the discriminative feature information from the different convolutional layers, we propose a CFFM, the structure of which is shown in Fig. 4. Note that the advanced semantic information from the deep layers can help improve the performance of the classification network, the last layers from Conv5_1 and Conv5_3 are fused.
The specific process of CFFM is as follows:
-
1.
Generation of feature maps Z1 and Z2: First, a 1 × 1 convolution operation is used for the last feature layers of the Conv5_1 and Conv5_3 modules from the backbone network respectively, resulting in two feature maps Z1 ∈ RC×W×H and Z2 ∈ RC×W×H, where C is the number of channels, W and H are the width and height respectively, followed by a 1 × 1 convolution operation for increasing information flow among channels. In this paper, C is set to 2048, the dimensions of Z1 and Z2 are the same as those of the outputs of the last layers from Conv5_1 and Conv5_3, respectively.
-
2.
Multi-level feature space fusion: the feature map Z3 is obtained by the cross-layer fusion of feature maps Z1 and Z2, and its calculation formula is as follows (1); then the Z3 is fed into the global average pooling (GAP) layer to attain FP ∈ RC×1×1; finally, it is fed into the fully connected layer to obtain a classification result.
$$Z_{3} = Z_{1} \otimes Z_{2}$$(1)where Z1 ∈ RC×W×H, Z2 ∈ RC×W×H, Z3 ∈ RC×W×H, \(\otimes\) stands for pixel-wise multiplication.
Feature enhancement module based on down-sampling
As we know, CNN can extract rich feature information from feature maps, and then this feature information can be used to classify objects by the classification network. For fine-grained image classification tasks such as classification of normal and tumor cells, the ability of using CNN to obtain discriminative features is limited due to the great similarity among input images. Note that, ECANet25 is an improvement upon SENet32, designed to avoid the negative effects of feature compression and enhance cross-channel information extraction. ECANet is introduced into our network to enhance discriminative information extraction. Given the fact that different channels contain different semantic information in a feature map, the importance of extracted features from each channel is different, and even some extracted features may interfere with classification results. As a result, we use ECANet to screen out the discriminative features of cells more effectively and make the classification results more accurate.
The specific process of the ECANet algorithm is as follows: the original feature map F ∈ RC×W×H is fed into the GAP layer to extract the global feature Fp; followed by a k × k convolution for extracting cross-channel information Fe from Fp, where k is calculated by (2), γ and b are 2 and 1 respectively, and odd represents the nearest odd number; finally, Fe is fed into an activation function to obtain a result that is multiplied by the original feature map, thereby adjusting the channel relationship.
With the deepening of the network, the receptive fields of the deep layers become larger, but multiple down-sampling operations also give rise to the loss of the detailed features of cells. Therefore, in order to improve the extraction performance of the network, we enhance the features after each down-sampling. The structure of the feature enhancement module based on down-sampling is shown in Fig. 5, where Convk represents the k-th stage of the classification network.
Loss function optimization
The softmax loss function, which is defined as follows, is often used in training a deep learning model and makes it achieve good classification accuracy.
However, it is noting that there exists a class imbalance between normal and tumor cells for the classification of normal and tumor cells. Besides, the morphological characteristics of normal and tumor cells are similar, and only the local characteristics are slightly different, which leads to low classification accuracy. To solve the above problems, we improve the softmax loss function, called w-softmax loss function. As we all know, the softmax loss function consists of the cross-entropy loss function and the softmax function. Next, the cross-entropy loss function is first introduced and defined as:
where M is the number of classes in the dataset, i expresses the i-th sample, pic represents the probability of i being class c, and yic is used to quantify the similarity between the predicted label and the true label c. Concretely, yic = 1 means exactly the same, while yic = 0 indicates completely different.
In this paper, only normal and tumor cells of white blood cells are classified. Therefore, with M = 2 and yi1 + yi2 = 1, Eq. (4) can be transform into Eq. (5), which represents the binary cross-entropy loss (BCELoss) function.
The number of normal cells is usually different from the number of tumor cells, leading to class imbalance. Therefore, in this paper, we improve the BCELoss by weighting, namely the losses of both classes are multiplied by weights to make the network model pay more attention to the smaller class in the training set. The improved BCELoss is given by:
where \(\alpha_{1}\) is the proportion of normal cells to all white blood cells in the training set. Similarly, \(\alpha_{2}\) is the proportion of tumor cells to all white blood cells in the training set. Finally, (3) and (6) are combined to attain the improved BCELoss as follows:
where \(\theta_{i}\) corresponds to the i-th column of the output matrix of the fully connected layer. \(x^{(i)}\) represents the characteristics of the input of the last layer of the fully connected layer, and bj represents the bias.
For the classification of normal and tumor cells, there are not only great similarities between them, but also differences in local characteristics. Owing to the above problems, for cell classification, it is highly recommended to maximize the inter-class separation and enhance the intra-class compactness to improve the classification accuracy. However, using the softmax loss in the classification network can only enforce the inter-class separation, but cannot enhance the intra-class clustering. To address this issue, we incorporate a center loss function33 in the training of the network. In other words, a constraint is added to the softmax loss function. The center loss function helps learn the feature centers of different cell classes and enhance the intra-class compactness. The center loss function is defined as:
where xi is the extracted feature map of the i-th image. \(c_{{y_{i} }}\) represents the center of the \(y_{i}\)-th cluster, and m is the number of images in a mini-batch. Ideally, the center of each cluster should be updated every epoch, but this is too inefficient. Therefore, in the training process, the center of each cluster is updated based on the mini-batch m, and the partial derivative of \({\mathcal{L}}_{C}\) with respect to xi and the update gradient of \(c_{{y_{i} }}\) are shown as.
where \(\delta (condition)\) = 1 if the condition is satisfied, otherwise \(\delta (condition)\) = 0.
In this paper, the improved BCELoss function and the center loss function are combined. The former can ensure the feature separation between classes, meanwhile also effectively improve the decline of classification accuracy caused by class imbalance. The latter can make images from the same class cluster. In other words, this approach can enhance the intra-class compactness.
The combination of these two loss functions (7) and (8) improves network performance. Which is defined as:
where m is the number of images in a mini-batch, \(\lambda\) and \(\beta\) are the weight coefficients, which are used to adjust the proportion of the two parts and set to 1 and 0.05, respectively.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Experiments and results
Dataset
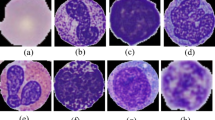
ALL-IDB34, which consists of 108 images and contains about 39,000 blood elements, and each image resolution is 2592 × 1944, is used to study the segmentation and classification of white blood cells. The BCCD dataset35 includes 364 microscopic images of various white blood cells. In ALL-IDB, however, only 260 cytopathological cells can be used for classification. For classification tasks, the number of cytopathological images in the dataset is too small, which will seriously affect the network performance. Therefore, in this paper, we use a dataset with more cytopathological images, namely the C-NMC 2019 dataset36. This was released in the Medical Imaging Challenge of the International Symposium on Biomedical Imaging (ISBI). We use a subset of the C-NMC 2019 dataset, which contains white blood cell images from 118 patients. There are 12,528 white blood cells in the dataset, including 8491 cancer cells and 4037 normal cells, each cell of which is annotated by pathologists. Each image resolution is 2560 × 1920, and each blood smear contains multiple cells. To better classify white blood cells, the researchers segmented each white blood cell from the blood smear images to construct a dataset, in which each image resolution is 300 × 300. The dataset is classified into three subsets: a training set, a validation set and a testing set, and these three subsets are split with the ratio 7:1:2.
The white blood cells from the blood smears are shown in Fig. 6. Except for white blood cells, a blood smear contains other similar cells that may affect the classification performance. Therefore, the separated white blood cell images are adopted to train the classification network, which is expected to achieve better classification accuracy.
Configuration
In this paper, we implemented the classification method in Python on a computer with two NVIDIA 1080Ti GPUs, each with 11 GB of memory, and an Intel Xeon E5-2630 CPU. The pytorch library served as a high-level framework, and the experimental platform is the Ubuntu 18.04 system. The hyper-parameter details of the classification model are shown in Table 2. Note that we adopted the vanilla cross-entropy loss function to guide the training of both ResNeXt-50 and ResNeXt-101 (section "Effect of different depths on network performance"), while we employed the proposed w-softmax loss function for our model in other experiments (section "Effect of color models on network performance" to "Comparison with CNN-style models").
Performance evaluation metrics
In this paper, tumor cells are positive samples and normal cells are negative samples. The performance evaluation metrics can be described by four values:
-
1.
TP: true positives, the number of correctly classified tumor cells.
-
2.
FP: false positives, the number of normal cells classified as tumor cells.
-
3.
TN: true negatives, the number of normal cells correctly classified.
-
4.
FN: false positives, the number of tumor cells classified as normal cells.
Currently, the most widely used metrics for the quantitative evaluation of image classification results are the following: Accuracy, Precision, Recall, F1-score, Specificity and AUROC. Five of these are defined by:
The AUROC metric refers to the area under the receiver operating characteristic (ROC) curve.
Ablation experiment
Effect of different depths on network performance
In CNN, simply increasing the network depth does not necessarily improve its performance. To validate the effect of the network depth on the classification performance, we conducted experimental research on ResNeXt network with different depths using the HSV color space. The experimental results are shown in Table 3.
From Table 3, the deeper network layers don’t mean the better performance in medical image domain. When the network deepens to a certain depth, the network performance will decline, although the Specificity of ResNeXt-101 is higher. Figure 7 illustrates a ROC comparison between ResNeXt-50 and ResNeXt-101. It is evident that ResNeXt-50 outperforms ResNeXt-101 in cell classification, so we have chosen ResNeXt-50 as the backbone network for this paper.
Effect of color models on network performance
In this section, we explore the effect of color space transformation on cell classification. As a common color space model, the RGB color space is widely used in medical imaging domain. However, for different applications we should select the most suitable color space. Specifically, we investigated RGB and HSV color spaces in terms of classification accuracy.
It can be seen from the experimental results in Table 4 that the classification accuracy is improved after transforming images from the RGB color space into the HSV color space, which proves that cytopathological images in the HSV color space can express image features better than those in the RGB color space. Cytopathological cell images in the HSV color space can better highlight local features and increase the contrast between cell classes. In contrast to the RGB color space, the three metrics of Accuracy, F1-score, AUROC and Specificity of the HSV color space are 0.16%, 0.42%, 0.07% and 1.24% higher, respectively. For the above reasons, we choose HSV as our color space model for white blood cell classification.
Effect of feature fusion on network performance
In Table 5, we show the effect of CFFM on our network. Notably, our experiments are based on the HSV color model. When the last layer of Conv5_3 fuses with the last layers of Conv5_1 and Conv5_2, respectively, all metrics of the classification network are improved. The accuracy, F1-score, AUROC, and specificity of the Conv5_1 + Conv5_3 method is 0.59%, 0.43%, 0.53%, and 1.08% higher, respectively, than those of the Conv5_2 + Conv5_3 method.
It proves that convolutions on different levels contain different discriminative information, and for the fine-grained classification task, local features play an important role in distinguishing different classes. Thus, the CFFM proposed in this paper can better obtain the discriminative information of cells by fusing different level features. In contrast, we choose the feature fusion of Conv5_1 and Conv5_3.
Effect of composite loss function on network performance
In this section, our experiments are based on both the HSV transformation and CFFM. To further boost the cell classification performance, we improve the softmax loss function (w-softmax loss function) and combine the w-softmax loss function with the center loss function to achieve better results. In Table 6, we compare the results of softmax loss, w-softmax loss and w-softmax loss + center loss. From the comparison of Table 6, it is easily observed that, when using our loss function, the Accuracy, F1-score, AUROC and Specificity reach optimal performance, 87.46%, 90.62%, 93.87% and 77.72%, respectively. Compared with the original softmax loss, our network performance is slightly increased by the w-softmax loss, and our loss function is the most competitive since it can maxmize the inter-class separation and improve the intra-class clustering.
Effect of feature enhancement module based on down-sampling on network performance
In this section, our experiments are based on the HSV transformation, CFFM and composite loss function. For fine-grained classification of normal and tumor cells, it is important to enhance the discriminative ability of cell details. However, not all extracted cell features are useful for the classification task. Meanwhile, some important information may be lost in the down-sampling process. Therefore, we constructed a down-sampling feature enhancement module that introduces ECANet into the down-sampling process to enhance the network's discrimination for useful features.
In Table 7, we compared the white blood cell classification performance between using down-sampling (ECANet) and pooling. The experiment shows that by placing the attention module in the down-sampling layer, the loss of useful information of images is mitigated and the discriminative information of images is extracted more efficiently, and thus dramatically improving the network performance. In contrast to not using ECANet, the four metrics of Accuracy, F1-score, AUROC and Specificity are 1.99%, 1.46%, 0.86% and 3.39%, respectively. Figure 8 visualizes the feature heatmaps from the last convolutional layer of the network, which are drawn using the Grad-CAM algorithm31. In the heatmap, the redder area represents the greater response of the network to that area. That is to say, the redder area represents more attention being rendered. On the contrary, the bluer area represents less attention being rendered. Clearly, with the addition of the attention module (ECANet), the network responds more to cells, particularly emphasizing their edges and content.
Comparison with CNN-style models
To validate our network performance, in this section, we compare our algorithm with CNN-style models. The classification results are shown in Table 8.
From the results in Table 8, ResNet-50 has achieved good results in cell classification. Furthermore, both ResNet-50 and our method have adopted the residual connection, which further proves that the residual connection method used in our algorithm is helpful to improve the classification accuracy. Densenet121 uses dense blocks to extract image features, because the network is too complex, thereby leading to over-fitting. That is to say, the classification results are not very good. In a word, the Accuracy, F1-score, AUROC and Specificity of our method are 8.57–4.66%, 6.86–3.21% and 7.26–3.19% and 11.68–6.12% higher than those of other models, respectively. The comparison of the ROC of the above classification algorithms is shown in Fig. 9, from which we can see that our method has a clear advantage over the other methods. To sum up, our method is superior to these common classification methods in various classification measures, which fully proves the superiority of our method.
Comparison with ViT-style models
To further validate our network performance, in this section, we compare our algorithm with ViT models. From Table. 9 we can observe the classification results. Compared with other ViT-style models, the Accuracy, F1-score, AUROC and Specificity of our algorithm are 10.39–20.43%, 13.7–23.56%, 13.1–25.71%, and 22.78–36.98% higher than those of the other models. These above results confirm that our method is significantly superior to the other five ViT-style models.
Besides, to more intuitively evaluate classification performance between the four ViT-style models and our method, we have constructed a bar chart. Figure 10 shows our method has an overall advantage.
Discussion
Regarding the diversity of training data, deep learning-based classification models necessitate a substantial corpus of high-quality annotated data to achieve robust performance. Nevertheless, acquiring such a comprehensive dataset can be challenging, particularly in scenarios where data availability is limited. Therefore, data augmentation techniques are potentially effective approaches. In this study, we employ traditional data augmentation methods, including horizontal flips, vertical flips, random scaling, and random rotation transformations, during the model training. However, conventional data augmentation technologies are limited in their ability to simulate real data diversity. To address this, more effective data augmentation and utilization methods should be introduced into our network. Specifically, future research could explore the application of semi-supervised learning and distillation techniques to mitigate the reliance on extensive annotated datasets. Additionally, incorporating generative adversarial networks (GANs) offers a promising avenue for further enriching our dataset.
In terms of attention mechanism in network architecture, the self-attention mechanism has demonstrated significant potential. The self-attention technique allows the model to focus on the most informative regions within the input, prioritizing the relevant features that contribute to the classification task. By effectively combining CNN and the self-attention mechanism, we can capture multi-scale contextual features from different network layers, thereby improving the accuracy of white blood cell classification. Moreover, building upon the self-attention technique, the image-content-based biomarkers of white blood cells can be extracted through interpretable visualized analysis of the white blood cell classification process. This enables a deeper understanding of the underlying features and patterns that the model has learned, potentially leading to new insights and the development of more robust and clinically relevant classification systems.
Conclusion
In summary, while the morphological characteristics of normal and tumor white blood cells are quite similar, they exhibit subtle differences in localized areas. Drawing inspiration from the ResNeXt classification network, we proposed a ResNeXt-CC classification network for white blood cells. Our method offers several advantages: (1) the proposed CFFM enhances the ability of discriminative information extraction; (2) a feature enhancement module, based on down-sampling, strengthens cell detail extraction; (3) the combination of w-softmax loss and center loss improves intra-class compactness and ensures inter-class separation; (4) color space transformation is applied to better represent cell texture information. The experimental results on the C-NMC 2019 dataset show that our proposed method manifests obvious advantages over the existing classification methods, including ResNet-50, Inception-V3, Densenet121, VGG16, Cross ViT, Token-to-Token ViT, Deep ViT, and simple ViT about 5.5–20.43% accuracy, 3.6–23.56% F1-score, 3.5–25.71% AUROC and 8.1–36.98% specificity, respectively. Ultimately, we hope that our work will inspire other related research on white blood cell classification.
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ferlay, J., et al. Global Cancer Observatory: Cancer Today (International Agency for Research on Cancer, Lyon, France, 2020). https://gco.iarc.fr/today.
Zheng, R. et al. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Center 2, 1–9 (2022).
Mohapatra, S. & Patra, D. Automated cell nucleus segmentation and acute leukemia detection in blood microscopic images. In 2010 International Conference on Systems in Medicine and Biology 49–54 (IEEE, 2010).
Sabino, D. M., Da Fontoura-Costa, L., Gil-Rizzatti, E. & Antonio-Zago, M. A texture approach to leukocyte recognition. Real-Time Imaging 10(4), 205–216 (2004).
Wijesinghe, C. B., Wickramarachchi, D. N., Kalupahana, I. N., De Seram, L. R., Silva, I. D. & Nanayakkara, N. D. Fully automated detection and classification of white blood cells. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (2020).
Deshpande, N. M., et al. Improved Otsu and Kapur approach for white blood cells segmentation based on LebTLBO optimization for the detection of Leukemia. Math. Biosci. Eng. 19(2), 1970–2001 (2022).
Rokach, L. & Maimon, O. Clustering methods. In Data Mining and Knowledge Discovery Handbook 321–352 (Springer, 2005).
Ruspini, E. H. A new approach to clustering. Inf. Control 15(1), 22–32 (1969).
Xu, R. & Wunsch, D. Clustering Vol. 10 (Wiley, New York, 2008).
Lippeveld, M. et al. Classification of human white blood cells using machine learning for stain-free imaging flow cytometry. Cytometry Part A 97(3), 308–319 (2020).
Zheng, A. & Casari, A. Feature Engineering for Machine Learning: Principles and Techniques for Data Scientists (O’Reilly Media Inc, Sebastopol, 2018).
Toğaçar, M., Ergen, B. & Cömert, Z. Classification of white blood cells using deep features obtained from convolutional neural network models based on the combination of feature selection methods. Appl. Soft Comput. 97, 106810 (2020).
Hung, J. & Carpenter, A. Applying faster R-CNN for object detection on malaria images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 56–61 (2017)
Duggal, R., et al. SD-layer: stain deconvolutional layer for CNNs in medical microscopic imaging. In International Conference on Medical Image Computing and Computer-Assisted Intervention 435–443 (Springer, Cham, 2017).
Shahin, A. I. et al. White blood cells identification system based on convolutional deep neural learning networks. Comput. Methods Progr. Biomed. 168, 69–80 (2019).
Huang, P. et al. Attention-aware residual network based manifold learning for white blood cells classification. IEEE J. Biomed. Health Inform. 25(4), 1206–1214 (2020).
Shi, J. et al. Cervical cell classification with graph convolutional network. Comput. Methods Progr. Biomed. 198, 105807 (2021).
Ha, Y., Du, Z. & Tian, J. Fine-grained interactive attention learning for semi-supervised white blood cell classification. Biomed. Signal Process. Control 75, 103611 (2022).
Sharma S, Gupta S, Gupta D, et al. Deep learning model for the automatic classification of white blood cells[J]. Computational Intelligence and Neuroscience, 2022, 2022.
Zheng, X. et al. White blood cell detection using saliency detection and CenterNet: A two-stage approach. J. Biophoton. 16(3), e202200174 (2023).
Tummala, S. & Suresh, A. K. Few-shot learning using explainable Siamese twin network for the automated classification of blood cells. Med. Biol. Eng. Comput. 61(6), 1549–1563 (2023).
Dhalla, S., Mittal, A. & Gupta, S. LeukoCapsNet: a resource-efficient modified CapsNet model to identify leukemia from blood smear images. Neural Comput. Appl. 36(5), 2507–2524 (2024).
Gao, S., et al. GAN-assisted sample equalization for CNN-based human peripheral leukocyte image recognition and classification. In 2024 7th World Conference on Computing and Communication Technologies (WCCCT) 324–329 (IEEE, 2024).
Xie, S., et al. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 1492–1500 (2017).
Wang, Q., et al. ECA-Net: Efficient channel attention for deep convolutional neural networks. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2020).
He, K., et al. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 770–778 (2016).
Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision [C]. IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, USA, 2016: 2818–26.
Lin, T. Y., et al. Bilinear cnn models for fine-grained visual recognition. In Proceedings of the IEEE International Conference on Computer Vision 1449–1457 (2015).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18 234–241 (Springer, 2015).
Lin, T. Y., et al. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2117–2125 (2017).
Selvaraju, R. R., et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision 618–626 (2017).
Hu, J., Shen, L. & Sun G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 7132–7141 (2018).
Wen, Y., et al. A discriminative feature learning approach for deep face recognition. In European Conference on Computer Vision 499–515 (Springer, Cham, 2016).
Labati, R. D., Piuri, V. & Scotti, F. All-IDB: The acute lymphoblastic leukemia image database for image processing. In 2011 18th IEEE International Conference on Image Processing 2045–2048 (IEEE, 2011).
BCCD, Blood Cell Images. www.kaggle.com/datasets/paul-timothymooney/blood-cells. Accessed 25 Sept 2020.
Gupta, A., et al. Classification of normal vs malignant cells in B-ALL white blood cancer microscopic images. In IEEE International Symposium on Biomedical imaging (ISBI)-2019 Challenges Internet (2019).
Huang, G., et al. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 4700–4708 (2017).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
Chen, C. F. R., Fan, Q. & Panda, R. Crossvit: Cross-attention multi-scale vision transformer for image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision 357–366 (2021).
Yuan, L., et al. Tokens-to-token vit: Training vision transformers from scratch on imagenet. In Proceedings of the IEEE/CVF International Conference on Computer Vision 558–567 (2021).
Zhou, D., et al. Deepvit: Towards deeper vision transformer. arXiv preprint arXiv:2103.11886 (2021).
Beyer, L., Zhai, X. & Kolesnikov, A. Better plain vit baselines for imagenet-1k. arXiv preprint arXiv:2205.01580 (2022).
Funding
The research reported here was, in part, supported by Doctoral Scientific Research Foundation of Anshan Normal University (No. 22b08), and Department of education in Liaoning Province China (No. LJKMZ20221811). This work is also supported by the 14th Five-Year Plan Special Research Project of Anshan Normal University (No. sszx013).
Author information
Authors and Affiliations
Contributions
Y.L. designed the algorithm model and wrote the main manuscript text, Y.X. and C.W. prepared and pre-processed the white blood cell image data, Q.L. investigated related researchers and design the program, C.F. provided the computing resource and validated the experimental results, and H.J. revised and verified the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, Y., Xu, Y., Wang, C. et al. ResNeXt-CC: a novel network based on cross-layer deep-feature fusion for white blood cell classification. Sci Rep 14, 18439 (2024). https://doi.org/10.1038/s41598-024-69076-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69076-1
Keywords
This article is cited by
-
WeedSwin hierarchical vision transformer with SAM-2 for multi-stage weed detection and classification
Scientific Reports (2025)
-
A two stage blood cell detection and classification algorithm based on improved YOLOv7 and EfficientNetv2
Scientific Reports (2025)