Abstract
Sepsis, a life-threatening syndrome, continues to be a significant public health issue worldwide. Sialylation is a hot potential marker that affects the surface of a variety of cells. However, the role of genes related to sialylation and sepsis has not been fully explored. Bulk RNA-seq data sets (GSE66099 and GSE65682) were obtained from the open-access databases GEO. The classification of sepsis samples into subtypes was achieved by employing the R package “ConsensusClusterPlus” on the bulk RNA-seq data. Hub genes were discerned through the application of the R package “limma” and univariate regression analysis, with the calculation of risk scores carried out using the R package “survminer”. To identify the best learning method and construct a prognostic model, we used 21 different combinations of machine learning, and C-index ranking results of these combinations have been showed. ROC curves, time-dependent ROC curves, and Kaplan–Meier curves were utilized to evaluate the diagnostic accuracy of the model. The R packages “ESTIMATE” and “GSVA” were employed to quantify the fractions of immune cell infiltration in each sample. The bulk RNA-seq samples were categorized into two distinct sepsis subtypes utilizing 14 prognosis-related sialylation genes. A total of 20 differentially expressed genes (DEGs) were identified as being associated with the relationship between sepsis and sialylation. The RSF was used to identify key genes with importance scores higher than 0.01. The nine hub genes (SLA2A1, TMCC2, TFRC, RHAG, FKBP1B, KLF1, PILRA, ARL4A, and GYPA) with the importance values greater than 0.01 was selected for constructing the prognostic model. This research offers some understanding of the relationship between sepsis and sialylation. Besides, it contains one predictive model that might develop into diagnostic biomarkers for sepsis.
Similar content being viewed by others
Introduction
Sepsis arises from multiple organ system dysfunction, which is often caused by a dysregulated host response to infection1. Characterized by high morbidity and mortality, sepsis can also cause irreversible physical damage2. Additionally, when sepsis progresses to septic shock, the mortality rate of patients in the intensive care unit (ICU) could reach as high as 38%3. At present, development in medicine and a better understanding of sepsis have improved the early diagnosis and treatment of sepsis, leading to a gradual decrease in mortality rate4. However, we still lack a comprehensive understanding of the pathogenesis of sepsis, which requires in-depth research to further reduce its incidence and mortality rate.
The process of sialylation plays a vital and essential role during infections, particularly in those affecting the respiratory tract. Several kinds of viral proteins undergo terminal glycosylation when viruses invade the human body, but this process could be effectively disrupted utilizing substances that inhibit sialidase such as neuraminidase inhibitors, so as to suppress to the infections5. Sialic acid is involved in the structure of transmembrane receptors on the surface of immune cells and plays a critical role in immune balance during sepsis immune dysfunction by modulating inflammation and immunity with a bi-directional effect, macrophage polarization, and suppressing neutrophil function during infection6. These findings indicated that sialylation-related genes (SRGs) can serve as a valuable indicator during the inflammatory process, providing insights into the activity of the immune system. Moreover, a decrease in sialylation level may signify the elimination of inflammation and regulation and recovery of the immune system.
Previous research has predominantly focused on examining the expression of genes in the peripheral blood of sepsis patients, aiming to elucidate the molecular mechanisms underlying these differential genes7. However, there were currently no relevant studies probing into the pathogenesis of sepsis from the aspect of combining the mechanisms of sepsis and sialylation, and potential diagnostic or therapeutic targets related to salivation remained unclear. In this study, we identified genes associated with both sepsis and sialylation. Subsequently, using the random survival forest (RSF) machine learning, we developed a prognostic model to improve the diagnosis of sepsis. The present discoveries provided valuable insights for the prognostic assessments and development of treatment strategies for sepsis.
Materials and methods
Data sources and processing
Blood samples from the GSE66099 dataset (containing 276 samples, with 47 healthy and 199 sepsis cases) and the GSE65682 dataset (containing 802 samples, with 42 healthy and 760 sepsis cases) were obtained from the Gene Expression Omnibus (GEO) database. Based on the two datasets, the probe expression matrix was downloaded after normalization and log2 transformation. The annotation file was then retrieved from the detection platform to map the gene symbol to the probe number. When probes mapped more than one symbol were removed, but if different probes were mapped to the same gene symbol, the mean of the probes was used taken as the final expression value for the gene. These samples were processed as a bulk RNA-seq dataset and batch effects were corrected and the data were merged using the R packages “limma” and “sva”. The latest sialylation-related genes (SRGs) including sialyltransferases, transporters, and neuraminidases were identified based on the Molecular Signatures Database (MSigDB)8.
Consensus clustering analysis
Univariate Cox regression analysis on the complete sample set was performed to identify prognostic genes with a p-value < 0.05. Next, consensus cluster analysis was conducted using "ConsensusClusterPlus" in the R package. We repeated 1000 iterations to ensure the stability of the clustering results and considered a range of cluster numbers from 2 to 9. The clustering stability threshold was set to 0.8, meaning that the number of clusters was valid only if the consistency index of the clustering results was greater than or equal to this value. In each iteration, clustering was performed with a random sampling proportion of 80% of the samples, and the distance was measured using the hierarchical clustering algorithm and Pearson's correlation coefficient. The optimal number of clusters was determined according to cumulative distribution function (CDF) and Delta area plot, and we identified biologically characterized molecular clusters.
Subsequently, KM survival curves were generated using the R package "survminer" and the log rank test was applied to evaluate prognostic differences among the identified subtypes. To investigate the factors influencing the prognostic differences among the distinct subtypes, the data on HALLMARK, Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.kegg.jp/kegg/kegg1.html), and Reactome pathways from the MSigDB database were collected9,10,11. The R package "GSVA" was employed to analyze the variations in the pathway activity between the subtypes and the variances were visualized into heatmaps using the R package "pheatmap".
Differential analysis and functional enrichment analysis
Using the 'limma' package, differential analysis was performed on various groups, including the subtypes identified by the consensus clustering analysis, healthy samples, and sepsis samples. Genes that met the thresholds of |log2 (fold-change) |> 1 and p-value < 0.05 were considered as differentially expressed genes (DEGs), which were visualized into volcano plots or heatmaps using the R package 'ggplot2'. The DEGs were subjected to functional enrichment analysis of KEGG and Gene Ontology (GO) analysis using the R package 'clusterProfiler'.
Establishment and validation of a prognostic model based on the prognostic genes in sepsis
Univariate Cox regression analysis was performed on the DEGs between two subtypes to select genes with a p-value < 0.001 as prognostic genes. Visualization of the results into forest plot was achieved using the R package “survminer”. Subsequently, these prognostic genes were intersected with the DEGs identified between the normal and sepsis samples. To identify the optimal learning method and to construct a prognostic model, we used 21 different combinations of machine learning algorithms and ranked the results using C-index. Finally, the RSF, which showed the highest C-index, was used to identify key genes with importance scores higher than 0.01 to develop a prognostic model. In addition, ROC curves, time-dependent ROC curves, and KM curves plot were used to assess the diagnostic performance of the model.
Analysis of immune cell infiltration
The workflow chart of this study is shown in Supplementary Fig. 1. To analyze the immune cell characteristics, immune cell infiltration fractions in each sample were quantified using the R package “ESTIMATE” and the ssGSEA method in the R package “GSVA”. Differences in the expression of immune-infiltrating cells between normal and sepsis samples and the correlation between each hub gene and a type of immune-infiltrating cell were visualized by “ggplot2”.
Results
Screening of key sialylation-related genes in sepsis
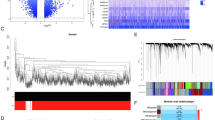
After removing batch-effects between the samples, RNA-seq data in sepsis from GSE65682 and GSE66099 were integrated for subsequent analysis and principle component analysis (PCA) plots were generated to show the distribution of samples (Fig. 1A,B). Combined with the prognostic data of the patients, we performed univariate COX regression analysis and correlation analysis for 62 sialylation-related genes in RNA-seq expression profile. Among them, CMAS, GCNT1, GALNT10, NEU4, B3GNT3, B4GALNT1 were considered as risk factors for the prognosis of patients with sepsis, while PSAP, GALNT11, GNS, FCN1, SIGLEC7, ST3GAL6, NEU1, ST6GALNAC2 were considered as the favorable factors for the prognosis of sepsis (Fig. 1C). Based on the expression of these prognostic genes, sepsis samples were divided into two subgroups (cluster-A and cluster-B) (Fig. 2A, Supplementary Fig. 2). KM survival curve showed that the prognosis of cluster-B was significantly better than that of cluster-A (Fig. 2B). The box plot (Fig. 2C) showed that in group B, which had a better prognosis, the expression of the protective genes (GALNT10, NEU4, GCNT1, CMAS, B3GNT3, B4GALNT1) was significantly higher than that of group A, and that the expression of risk genes (SIGLEC7, FCN1, PSAP, GNS, ST6GALNAC2, NEU1, ST3GAL6) was significantly lower than that of group A. The distribution of the clinical characteristics of gender, age, survival status and time between the two subtypes was shown in Fig. 2D.
Analysis of Sialylation-related Genes in sepsis samples. (A,B) PCA plots of GSE65682 and GSE66099 datasets before (A) and after (B) integration by “limma” and “sva” packages. (C) Correlation analysis and Univariate regression analysis across 14 prognosis-related sialylation genes. Lines indicate a significant correlation between sialylation-related genes (p < 0.0001); purple indicates risk factors and green indicates favorable factors for OS.
Identification of two sepsis-subtypes by consensus clustering analysis based on prognosis-related sialylation genes. (A) Consensus matrix plots. K = 2 was determined as the optimal clustering number. (B) Kaplan–Meier survival analysis in clusters. (C) Differential expression of prognosis-related sialylation genes in two sepsis subtypes. (D) Heatmap of the interaction between prognosis-related sialylation genes and clinicopathological features in sepsis. (*p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001).
Functional enrichment analysis on different sepsis clusters
To explore the differences in biological function between the two sepsis clusters, we performed HALLMARK, KEGG, and Reactome pathway enrichment analyses. The HALLMARK pathway (Fig. 3A) enrichment analysis showed that cluster A was enriched in TNFA signaling via NFKB, IL6-JAK-STAT3 signaling and inflammatory response. For KEGG pathway (Fig. 3B), cluster A was involved in JAK-STAT signaling pathway, Natural Killer cell-mediated cytotoxicity and B cell receptor signaling pathway. In Reactome analysis (Fig. 3C), cluster A was enriched in interleukin 10 and 15 signaling pathway, CD28-dependent vav1 pathway and FCERL-mediated MAPK activation. The results of the above enrichment analysis showed that cluster A was closely related to immune regulation and inflammatory progression that could results in a poor prognosis of sepsis. PCA plot confirmed that there existed differences between the two sepsis clusters (Fig. 4A). To further explore different sepsis subgroups, we calculated 112 DEGs between the two clusters (Fig. 4B). The GO and KEGG enrichment results (Fig. 4C,D) demonstrated the distribution of these DEGs in different pathways and biological processes including immune and inflammation-related pathways, which were also cross-verified with the results of the previous analysis.
Functional enrichment analysis between two clusters by different database. (A) Heatmap results of HALLMARK pathway (B) KEGG pathway (www.kegg.jp/kegg/kegg1.html) (C) and Reactome pathway enrichment analysis.
Identification and analysis of the DEGs between healthy and sepsis samples
After normalizing the dataset (Fig. 5A,B), we identified a total of 8167 genes (4354 upregulated genes and 3813 downregulated genes) with significant expression differences between the healthy and the sepsis groups (Fig. 5C,D). GO and KEGG functional enrichment analysis was used to further explore these genes. GO results (Fig. 6A–C) showed that these genes played important roles in processes such as regulation of T cell activation, positive regulation of leukocyte cell–cell adhesion and nucleoside-triphosphatase regulator activity. KEGG pathway enrichment analysis results (Fig. 6D) displayed that T cell receptor signaling pathway, B cell receptor signaling pathway and Th17 cell differentiation were regulated by these DEGs.
Identification of DEGs between healthy and sepsis samples. (A,B) Homogenize the GSE65682 and GSE66099 datasets. Before homogenization (A); After homogenization (B). (C) The volcano map shows the distribution of DEGs between the healthy and sepsis samples. (D) The heat map shows the expression of the DEGs in healthy and sepsis samples.
Screening prognostic genes for sepsis
Univariate COX regression analysis was performed on the 112 DEGs between the two clusters. A total of 20 prognosis-related genes were screened with a p-value of < 0.001 (Fig. 7A). The intersection between 4354 upregulated DEGs and 3813 downregulated DEGs in the normal and sepsis samples contained a total of 20 genes (18 upregulated and 2 downregulated genes) (Fig. 7B,C). GO and KEGG function enrichment analysis was further performed on these 20 genes. GO analysis results showed that these genes were involved in response to oxygen levels, vascular transport and carbonate dehydratase activity (Fig. 7D). In KEGG analysis results, these genes were enriched in HIF-1 signaling pathway, insulin secretion and estrogen signaling pathway (Fig. 7E). The expression of these 20 genes was also shown in the form of volcano map and heatmap, in which KRT23 and PIRA were high-expressed in normal samples, while MS4A3, CD24, CEACAM8, TFRC, RAP1GAP, TMCC2, SLC2A1, HMBS, KLF1, FKBP1B, CA1, GYPB, CA2, RNF14, SLC7A5, ARL4A, GYPA, RHAG were highly expressed in sepsis samples (Fig. 8A,B).
Screening of prognostic-related genes in sepsis. (A) Univariate regression analysis was performed on the 112 DEGs between cluster -A and -B in sepsis samples. Forest plot shows 20 genes that meet the p-value < 0.001 criteria. (B) The intersection of the up-regulation DEGs with 20 genes. (C) The intersection of the down-regulation DEGs with 20 genes. GO (D) /KEGG (E) enrichment analysis of intersecting genes.
Construction of a prognostic model for sepsis
A variety of machine learning algorithms were used to build the predictive models. The C-index ranking results of these machine learning algorithms showed that the RSF, which had the highest C-index, was the most suitable for the screening of the model genes (Fig. 9A). We ranked genes based on their importance (Fig. 9B) and identified nine hub genes (SLA2A1, TMCC2, TFRC, RHAG, FKBP1B, KLF1, PILRA, ARL4A, and GYPA) with an importance value greater than 0.01. To explore the potential of these hub genes as diagnostic markers for sepsis, KM curves were plotted according to the overall expression of these 9 genes, and the results showed that a higher expression of these 9 genes was relate to a worse prognosis of sepsis patients (Fig. 9C). The AUC values for the ROC curves were 0.963 on the 1st day, 0.973 on the 14th day, and 0.979 on the 27th day. Both the time-dependent ROC curve and the normal ROC curves exhibited a high accuracy with AUC > 0.9, indicating the predictive capabilities of these genes (Fig. 9D,E).
Build prognostic model based on a variety of machine learning. (A) C-Index of 31 machine learning algorithms. (B) Random forest method result. (C) Survival analysis of machine learning score by random forest method. (D) Time-dependent ROC curve based on random forest method. (E) ROC curve based on random forest method.
Immune microenvironment analysis
The relationship between the immune microenvironment and sepsis was analyzed. Figure 10A displayed the correlation between 23 different types of immune cells in whole samples. Noticeably, there was a significant positive correlation between T follicular helper cells and monocytes, natural killer T cells. There was also a strong positive correlation between neutrophils and eosinophils, macrophages. Compared to normal samples, sepsis had lower levels of activated B cells, activated CD8 T cells, and immature B cells but significantly higher expression of activated dendritic cells, neutrophils, and type 17 T helper cells (Fig. 10B). Additionally, the extent of immune cell infiltration was evaluated. The expression of ARL4A, FKBP1B, GYPA, KLF1, RHAG, SLC3A1, TFRC, and TMCC2 was strongly correlated with type 17 T helper cells and CD56bright natural killer cells. Only PILRA was significantly associated with MDSCs, natural killer T cells, monocytes, and T follicular helper cells (Fig. 10C).
Assessment of the immune microenvironment. (A) Correlation between immune cells in the whole sample. (B) Differences in immune cells between healthy and sepsis samples. (C) Correlation of core genes (ARL4A, FKBP1B, GYPA, KLF1, PILRA, RHAG, SLC2A1, TFRC, TMCC2) in the prognostic model with immune cells.
Discussion
Antibodies play a vital role in bridging the adaptive and innate immune systems, and all human antibody classes fulfill biological functions through post-translational modifications of sugars12. Recent advances in glycomics showed that compared to IgG autoantibodies, IgG Fc N-glycosylation is a more reliable biomarker for the diagnosis and monitoring of a variety of autoimmune diseases through primarily modulating Fc domain conformation and Fcγ receptor binding affinity and signaling. Therefore IgG Fc N-glycosylation is considered as a potential therapeutic strategy to attenuate pathogenic autoimmunity13,14. Many of the glycan structures on the antibody are capped by sialic acid, and glycosylation plays an important role in the biology and function of all antibody classes, with a focus on terminal sialylation15. Studies have shown that lower levels of sialylated intrathecal IgG are associated with the onset of multiple sclerosis (MS)16. Sialylation of IgG-Fc is reduced in chronic inflammatory demyelinating polyneuropathy (CIDP), the level of which is correlated with clinical severity. The ratio of sialylation and sialylation/agaroylation of IgG-Fc may be a new means to monitor the severity of CIDP disease and treatment status17. In the Goodpasture mouse model, IgG sialylation effectively reduces disease severity by shifting antibody effector function to anti-inflammatory mode18. At the same time, patients with progressive rheumatoid arthritis have lower levels of IgG sialylation compared to those with milder or remission disease19. In addition to IgG, the role of sialylation in other antibody subclasses is gradually recognized. For example, sialylated IgE is associated with allergic pathogenicity, sialylated IgA has antiviral activity, and sialylated IgM could inhibit T cell proliferation15. For most types of antibodies, sialylation has a profound effect on the effector functions13.
Sepsis is a common complication of the combat against injury and trauma and is a life-threatening organ dysfunction caused by systemic inflammation and dysimmune response of the host to infection. Sepsis is also one of the significant causes of deaths in modern ICU20. Although the pathophysiology of sepsis remains elusive, immunosuppression is now considered as one of the leading causes to sepsis death. The systemic inflammatory response and deterioration of organ function in critically ill patients suggest a cascade of widespread overactivation of the host defense system21. For sepsis, traditional therapies may not be effective enough to prevent severe organ damage or death. Studies have shown that abnormal sialylation of carbohydrate structures on the cell surface affects host–pathogen interactions. α-2,6 salivary transferases (ST6GAL1) plays a key role in the immune response by altering the glycosylation pattern of host cell surface proteins infected by pathogens, thereby facilitating their invasion. Shipra Chandel et al. found a significant reduction in enteraggregative E. coli-induced MAPK, transcription factors, sialic glycoprotein, and IL-8 secretion in α-2,6 sialyltransferase-silenced cells when compared to the corresponding control cells22. Qingjie Fan et al. reported that ST6GAL1 deficiency can downregulate the expression of NF-κB and reduce the production of pro-inflammatory cytokines, thereby alleviating the onset of ulcerative colitis23. At present, though sialylation plays a critical role in controlling the production of new inflammatory cells and in inhibiting the cascade of inflammatory mediators in sepsis, there were no studies probing into the specific association between sialylation and the occurrence and progression of sepsis.
Extensive research has shown that a significant percentage of fatalities associated with sepsis can be attributed to uncontrolled opportunistic infections and the immunosuppressive traits that accompany the condition24,25. Sepsis has a direct effect on the immune system by causing adverse changes in how immune cells form, mature, function and death26. As we all know, Inflammatory cytokines such as IL-1β, IL-6 and TNF-α, NF-κB, MAPK and JNK signaling are activated in sepsis27,28. A study reported that knockdown of siglec-1 ((a member of sialic acid-binding Ig (I)-like lectin family members, the predominant sialic acid-binding proteins on cell surface) in RAW 264.7 cells resulted in inhibiting the production of TGF-β1 by ubiquitin-dependent degradation of Syk29. ST6Gal-I sialotransferase was reported to sialify TNFR1, thereby protecting tumor cells from TNF-induced apoptosis30, indicating sialylation could regulate inflammation. This study identified 2 clusters (cluster-A and cluster-B) for sepsis based on sialylation related genes, with cluster-A having a worse survival outcome. Our analysis results showed that Cluster A was mainly enriched in inflammation-related pathways (NF-κB, IL6-JAK-STAT3 signaling, TNF-α signaling, interleukin 10 and 15 signaling pathways), adaptive immune responses (JAK-STAT signaling pathway, natural killer cell-mediated cytotoxicity and B-cell receptor signaling pathways), as well as in classical cell signaling pathways (CD28-dependent vav1 pathway and FCERL-mediated MAPK activation).
A prognostic model incorporating 9 sialylation-related genes was developed and demonstrated strong performance for predicting sepsis survival outcome. Previous researches developed various signatures for predicting the prognosis and detection of sepsis. A highly predictive diagnostic model for sepsis detection was developed using 11 immune-related genes31. Based on ceRNA regulatory network analysis, a study identified four hub genes, including CD247, IL-1R2, IL-2Rβ and TGF-βR3 as novel biomarkers for evaluating the prognosis of sepsis32. Applying differential expression analysis, least absolute shrinkage and selection operator (LASSO) analysis, and multivariate logistic regression analysis, a four-gene (SIGLEC9, TSPO, CKS1B, and PTTG3P) risk prediction and evaluation model for sepsis and severe acute respiratory distress syndrome was established33. Another research established a robust prognostic model based on 22 DEIRGs to predict 28-day mortality and immunosuppression status for sepsis patients34. For the diagnosis of sepsis, a 25-gene signature diagnostic model has shown a high clinical value35. Noticeably, we were the first to conduct comprehensive bioinformatics analysis for sepsis based on sialylation-related genes, and we used fewer genes to predict the prognostic outcomes to facilitate the clinical translation.
In this study, we identified a total of 9 genes (ARL4A, FKBP1B, GYPA, KLF1, PILRA, RHAG, SLC2A1, TFRC, and TMCC2) associated with sialylation and sepsis prognosis. ADP ribosylation factor (Arf)-like 4A (ARL4A) is an Arf-small GTPase that plays a critical role in cell morphology, cell migration, and actin cytoskeletal remodeling36. However, the relationship between ARL4A and the occurrence and development of sepsis has not been clarified. FKBP1B belongs to the pro-immunolin superfamily, which is the cytosolic receptor protein of FK50637. It has been shown that improving FKBP1B can reverse calcium dysregulation and memory impairment in elderly rats, revealing the underlying molecular mechanism of brain aging and also the potential of using FKBP Prolyl Isomerase 1B (FKBP1B) as therapeutic target for Alzheimer's disease38. Glycophorins A (GYPA) is located on chromosome 4 and encodes the common MN blood group polymorphism antigen. GYPA gene evolves rapidly to evade the invasion of malaria parasites, but there are few studies probing into its correlation with the occurrence of common diseases39. Krüppel-like factor 1 (KLF1) is a transcription factor that regulates the expression of β-like globin genes, and any variation in the KLF1 gene may alter the phenotype of sickle cell disease40. Paired immunoglobulin-like type 2 receptor α (PILRA) plays a vital role in regulating a wide range of immune responses41. Weihua Shi et al. found that PILRA expression levels in rat models of atrial fibrillation are significantly higher than in rat models with sinus rhythm and are significantly positively correlated with the infiltration of multiple immune cells42. Zhongheng Zhang et al. used deep machine learning algorithms to construct a prognostic sepsis model that included PILRA, providing a theoretical support for the involvement of PILRA in regulating the occurrence and development of sepsis43. Human rh-associated glycoprotein (RHAG) is a transmembrane protein expressed only in red blood cells. RHAG binds to the Rh30 protein that carries the Rh blood group antigen and is expressed on the surface of red blood cells. One of the physiological roles of RHAG in erythrocytes is to regulate Rh30 membrane expression and maintain erythrocyte shape through several interactions44. Solute carrier family 2 member 1 (SLC2A1) is an important mediator of energy metabolism. Some studies have shown that exosomes in patients with sepsis targets SLC2A1 and inhibits glycolysis through has-mir-1262 and promotes apoptosis of human cardiomyocytes45. TFRC encodes transferrin receptor protein 1, which controls intracellular iron levels46. Xuebiao Wei et al. demonstrated that the exacerbation of sepsis-related encephalopathy may be caused by upregulated expression of TFRC to promote ferroptosis47. TMCC2 is an apoE-binding protein involved in the regulation of the development and progression of Alzheimer's disease by forming a complex with amyloid β protein precursors48. Although these genes have been previously reported, their association with sepsis was less studied.
However, there were still some limitations in the current study. This research was primarily based on bioinformatics analysis that lacked comprehensive clinical and experimental validation. Also, the correlation between sepsis and sialylation was still at the stage of ongoing research and development. It must be mentioned that the analysis has concerns about overfitting, since there is no validation of external independent datasets when available.
Conclusion
This study performed a comprehensive analysis to investigate the influence of sialylation in sepsis. Applying the RSF method, nine hub genes (SLA2A1, TMCC2, TFRC, RHAG, FKBP1B, KLF1, PILRA, ARL4A, and GYPA) that played crucial roles in the prognosis of sepsis were identified. This study improved the clinical diagnosis and treatment of sepsis.
Data availability
The datasets (GSE66099 and GSE65682) for healthy and sepsis samples were acquired from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) in this study.
References
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315(8), 801 (2016).
Schlapbach, L. J. Paediatric sepsis. Curr. Opin. Infect. Dis. 32(5), 497–504 (2019).
Liu, Y.-C. et al. Frequency and mortality of sepsis and septic shock in China: A systematic review and meta-analysis. BMC Infect. Dis. https://doi.org/10.1186/s12879-022-07543-8 (2022).
Gao, X.-M. et al. Identification of key genes in sepsis by WGCNA. Prev. Med. 172, 107540 (2023).
Pleass, R. J. The therapeutic potential of sialylated Fc domains of human IgG. mAbs https://doi.org/10.1080/19420862.2021.1953220 (2021).
Liu, Y.-C. et al. Sialic acids in the immune response during sepsis. Front. Immunol. https://doi.org/10.3389/fimmu.2017.01601 (2017).
Zhu, D., Zhu, K. & Guo, S. Identification of key genes related to immune cells in patients with gram-negative sepsis based on weighted gene co-expression network analysis. Ann. Transl. Med. 10(14), 787–787 (2022).
Zhou, M. et al. Characterization of sialylation-related long noncoding RNAs to develop a novel signature for predicting prognosis, immune landscape, and chemotherapy response in colorectal cancer. Front. Immunol. https://doi.org/10.3389/fimmu.2022.994874 (2022).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951 (2019).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587–D592 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Imbach, P. Treatment of immune thrombocytopenia with intravenous immunoglobulin and insights for other diseases. Swiss Med. Wkly. https://doi.org/10.4414/smw.2012.13593 (2012).
Li, D. et al. Sialylated immunoglobulin G: A promising diagnostic and therapeutic strategy for autoimmune diseases. Theranostics 11(11), 5430–5446 (2021).
Pincetic, A. et al. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 15(8), 707–716 (2014).
Vattepu, R., Sneed, S. L. & Anthony, R. M. Sialylation as an important regulator of antibody function. Front. Immunol. https://doi.org/10.3389/fimmu.2022.818736 (2022).
Kennedy, P. G. E. et al. Aberrant immunoglobulin G glycosylation in multiple sclerosis. J. Neuroimmune Pharmacol. 17(1–2), 218–227 (2021).
Wong, A. H. Y. et al. Sialylated IgG-Fc: A novel biomarker of chronic inflammatory demyelinating polyneuropathy. J. Neurol. Neurosurg. Psychiatry 87(3), 275–279 (2016).
Pagan, J. D., Kitaoka, M. & Anthony, R. M. Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell 172(3), 564-577.e13 (2018).
Engdahl, C. et al. Estrogen induces St6gal1 expression and increases IgG sialylation in mice and patients with rheumatoid arthritis: A potential explanation for the increased risk of rheumatoid arthritis in postmenopausal women. Arthr. Res. Ther. https://doi.org/10.1186/s13075-018-1586-z (2018).
Seymour, C. W. et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. Jama 321(20), 2003 (2019).
Liu, D. et al. Sepsis-induced immunosuppression: Mechanisms, diagnosis and current treatment options. Mil. Med. Res. https://doi.org/10.1186/s40779-022-00422-y (2022).
Chandel, S. et al. Role of ST6GAL1 and ST6GAL2 in subversion of cellular signaling during enteroaggregative Escherichia coli infection of human intestinal epithelial cell lines. Appl. Microbiol. Biotechnol. 107(4), 1405–1420 (2023).
Fan, Q. et al. Hyper α2,6-sialylation promotes CD4+ T-cell activation and induces the occurrence of ulcerative colitis. Adv. Sci. https://doi.org/10.1002/advs.202302607 (2023).
Akhmaltdinova, L. et al. Influence of pathogen type on neonatal sepsis biomarkers. Int. J. Inflam. 2021, 1009231 (2021).
Korzeniowski, T. et al. The role of the immune system in pediatric burns: A systematic review. J. Clin. Med. 11(8), 2262 (2022).
Zhang, W. Y. et al. Analysis and validation of diagnostic biomarkers and immune cell infiltration characteristics in pediatric sepsis by integrating bioinformatics and machine learning. World J. Pediatr. 19(11), 1094–1103 (2023).
Liu, X. et al. RIPK1 in the inflammatory response and sepsis: Recent advances, drug discovery and beyond. Front. Immunol. 14, 1114103 (2023).
Xian, L. et al. Systematic analysis of MASP-1 serves as a novel immune-related biomarker in sepsis and trauma followed by preliminary experimental validation. Front. Med. (Lausanne) 11, 1320811 (2024).
Wu, Y. et al. Induction of siglec-1 by endotoxin tolerance suppresses the innate immune response by promoting TGF-β1 production. J. Biol. Chem. 291(23), 12370–12382 (2016).
Holdbrooks, A. T., Britain, C. M. & Bellis, S. L. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J. Biol. Chem. 293(5), 1610–1622 (2018).
Xue, H. et al. A comprehensive analysis of immune features and construction of an immune gene diagnostic model for sepsis. BMC Genom. 24(1), 794 (2023).
Zhang, Q. et al. Screening of core genes prognostic for sepsis and construction of a ceRNA regulatory network. BMC Med. Genom. 16(1), 37 (2023).
Ming, T. et al. Integrated analysis of gene co-expression network and prediction model indicates immune-related roles of the identified biomarkers in sepsis and sepsis-induced acute respiratory distress syndrome. Front. Immunol. 13, 897390 (2022).
Zheng, Y. et al. Construction and validation of a robust prognostic model based on immune features in sepsis. Front. Immunol. 13, 994295 (2022).
Lai, Y. et al. Comprehensive analysis of molecular subtypes and hub genes of sepsis by gene expression profiles. Front. Genet. 13, 884762 (2022).
Chen, K.-J. et al. Arl4A and Pak1 cooperative recruitment to plasma membrane contributes to sustained Pak1 activation for cell migration. J. Cell Sci. https://doi.org/10.1242/jcs.233361 (2020).
Maalej, A. et al. Letter evidence of association between FKBP1B and thyroid autoimmune disorders in a large Tunisian family. Autoimmunity 37(3), 237–239 (2009).
Gant, J. C. et al. Reversal of aging-related neuronal Ca2+ dysregulation and cognitive impairment by delivery of a transgene encoding FK506-binding protein 12.6/1b to the hippocampus. J. Neurosci. 35(30), 10878–10887 (2015).
Naka, I. et al. The genotypes of GYPA and GYPB carrying the MNSs antigens are not associated with cerebral malaria. J. Hum. Genet. 52(5), 476–479 (2007).
Kumar, R. et al. Krüppel-like factor 1 (KLF1) gene single nucleotide polymorphisms in sickle cell disease and its association with disease-related morbidities. Ann. Hematol. 100(2), 365–373 (2021).
Li, Q. et al. Comprehensive analysis of PILRΑ’s association with the prognosis, tumor immune infiltration, and immunotherapy in pan-cancer. Sci. Rep. https://doi.org/10.1038/s41598-023-41649-6 (2023).
Shi, W. et al. PILRA is associated with immune cells infiltration in atrial fibrillation based on bioinformatics and experiment validation. Front. Cardiovasc. Med. https://doi.org/10.3389/fcvm.2023.1082015 (2023).
Zhang, Z. et al. Deep learning-based clustering robustly identified two classes of sepsis with both prognostic and predictive values. eBioMedicine 62, 103081 (2020).
Benjelloun, F. et al. Expression of the human erythroid Rh glycoprotein (RhAG) enhances both NH3 and NH4+ transport in HeLa cells. Pflügers Archiv Eur. J. Physiol. 450(3), 155–167 (2005).
Sun, F. et al. Exosomes derived from the blood of patients with sepsis regulate apoptosis and aerobic glycolysis in human myocardial cells via the hsa-miR-1262/SLC2A1 signaling pathway. Mol. Med. Rep. https://doi.org/10.3892/mmr.2022.12635 (2022).
Yi, L. et al. TFRC upregulation promotes ferroptosis in CVB3 infection via nucleus recruitment of Sp1. Cell Death Dis. https://doi.org/10.1038/s41419-022-05027-w (2022).
Wei, X.-B. et al. Exosome-derived lncRNA NEAT1 exacerbates sepsis-associated encephalopathy by promoting ferroptosis through regulating miR-9-5p/TFRC and GOT1 axis. Mol. Neurobiol. 59(3), 1954–1969 (2022).
Hopkins, P. C. R., Sáinz-Fuertes, R. & Lovestone, S. The impact of a novel apolipoprotein E and amyloid-β protein precursor-interacting protein on the production of amyloid-β. J. Alzheimer’s Dis. 26(2), 239–253 (2011).
Funding
This study was supported by the Key Social Development Project of Jiangsu Province (BE2021660), Suzhou Science and Technology Project (SKY2023197) and Clinical Research Project of Gusu School of Nanjing Medical University (GSKY20240202).
Author information
Authors and Affiliations
Contributions
Linfeng Tao and Lifang Wu conceived and designed the study. Linfeng Tao and Jun Liu collected data. Linfeng Tao and Yanyou Zhou wrote the paper. All the authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tao, L., Zhou, Y., Wu, L. et al. Comprehensive analysis of sialylation-related genes and construct the prognostic model in sepsis. Sci Rep 14, 18110 (2024). https://doi.org/10.1038/s41598-024-69185-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69185-x
Keywords
This article is cited by
-
Exploration of propionate metabolism-related genes to predict prognosis and immunotherapy response in ovarian cancer
Journal of Ovarian Research (2025)
-
Identification and validation of prognostic genes associated with m6A-regulated programmed cell death in acute lymphoblastic leukemia
Scientific Reports (2025)
-
Identification of Glycolysis-Related Signature and Molecular Subtypes in Child Sepsis Through Machine Learning and Consensus Clustering: Implications for Diagnosis and Therapeutics
Molecular Biotechnology (2025)
-
Comprehensive Analysis of Sialylation-Related Gene Profiles and Their Impact on the Immune Microenvironment in Periodontitis
Inflammation (2024)