Abstract
Marine and coastal ecosystems respond to climate change in various ways, such as the type of ecosystem, the species composition, interactions, and distribution, and the effect of local stressors. Metazoan organisms, particularly zooplankton, are important indicators for monitoring the effects climate-driven warming in marine coastal ecosystems over the long term. In this study, the diversity and distribution of zooplankton communities in the Mediterranean Sea (Canyon Dohrn and LTER-MareChiara, Gulf of Naples), a known biodiversity and climate changes hotspot, have been assessed using the integration of morphological-based identification and organismal eDNA. Our findings showed that the multi-locus strategy including the mitochondrial cytochrome c oxidase I (COI) gene and the hypervariable region V9 of the 18S rDNA (18S V9) as targets, improved the taxonomic overview, with the COI gene being more effective than the 18S V9 region for metazoans at the species level. However, appendicularians were detected only with the 18S V9 region. Overall, organismal eDNA is a powerful approach for revealing hidden biodiversity, especially for gelatinous and meroplankton components, and provided new insights into biodiversity patterns. The ecological importance of calanoid copepods in coastal ecosystems has been confirmed. In contrast, the discovery of 13 new metazoan records in the Mediterranean Sea, including two non-indigenous copepod species, suggested that local stressors affect zooplankton community structure and resilience, highlighting the importance of biomonitoring and protecting marine coastal ecosystems.
Similar content being viewed by others
Introduction
Marine metazoan zooplankton is a group of highly diverse multicellular organisms that either spend their entire life cycle in the water column (holoplankton) or temporarily live as microscopic planktonic life stages, occupying as adults the benthic or nekton environment (meroplankton). Zooplanktonic organisms perform critical ecological functions in marine ecosystems, linking primary producers (phytoplankton) to higher trophic levels (fish, seabirds, marine mammals) and regulating, directly and indirectly, ecosystem services such as carbon cycling and fish stocks1,2. Several studies have explored and modeled the zooplankton assemblages’ responses to climate-driven warming scenarios in marine ecosystems3,4,5,6. In this context, ecological data collected at long-term sites provide meaningful temporal scales baselines of biodiversity assessment, supporting the early identification of community changes, the plan and track of measures management, and the prediction and modeling of future relapses on the composition or function of marine ecosystems7,8,9,10.
The zooplankton taxonomic complexity, with ca. 2 × 105 species distributed in 11 different phyla11, has been traditionally identified and monitored by a microscopy-based approach, a time-consuming process requiring a high level of taxonomic expertise and almost impossible in the case of eggs, early larval live stages and gelatinous taxa with fragile bodies. Considering climate changes, it is essential to monitor the distribution and to identify possible shifts in the zooplankton communities in a relatively fast, cheap, and large-scale way. Over the past few decades, environmental DNA metabarcoding (eDNA) from the bulk samples (e.g., organisms collected using plankton nets or organisms isolated from sediment collected with sediment cores or grabs) or from the environmental matrix (e.g., water or sediment samples) has been reported to improve the limitation of morphology-based species identification and to be successful to uncover the marine metazoan diversity and to speed up environmental monitoring12,13,14. The non-invasive nature, cost-effectiveness, rapid turnaround, and high yield of data generated have made eDNA metabarcoding a very successful tool for assessing the diversity of marine zooplankton at spatial15,16 and/or temporal scales10,17,18,19. Such an approach has also been used to evaluate changes in vertical migrations of species and ecosystem responses to climatic events20 and to discover cryptic, non-indigenous, and rare species21,22. Despite its many advantages, there are several biases connected to the crucial choices needed for a successful application of eDNA. Among them, based on target taxa and the field of application, careful consideration is necessary for the selection of the substrate used as a source of the DNA sour (bulk samples or environmental matrix), the choice of primer pair (universal or specific), and the marker region, determining the further availability of reference sequences and the power discrimination at species level23,24,25.
This study aims to assess the diversity and distribution of the marine zooplankton metazoan community in the Gulf of Naples (Central Tyrrhenian Sea, Western Mediterranean Sea). A microscopy-based approach has been integrated with organismal eDNA (bulk samples) using two genetic markers: the mitochondrial cytochrome oxidase I (COI) gene and the hypervariable region V9 of the 18S ribosomal DNA (18S V9). The Mediterranean Basin is a global hotspot for biodiversity26 and climate change hotspot, with warming of this sea is expected to exceed global rates by 25%27,28. The Gulf of Naples, which spans roughly 195 km of coastline, is also highly affected by multiple anthropogenic pressures such as high population density, industrial and commercial activities, and tourism. In this study, we selected two different sites: the coastal site, LTER MareChiara (LTER-MC), and the Canyon Dohrn, located off-shore and about 12 nautical miles away. LTER MareChiara represents one of the most extended plankton time series available for the Mediterranean basin (https://www.st.nmfs.noaa.gov/copepod/time-series/it-30101/). The second site, Canyon Dohrn, is an active geological site is characterized by upwelling waters that provide crucial ecological benefits and serve as a breeding ground for various marine species along the coastline.
Results
General description
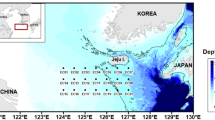
The data analyzed in this study were collected in two different sampling sites located in the Gulf of Naples. The first site is the coastal long-term ecological research site LTER MareChiara (LTER-MC), and the second site, Canyon Dorhn (C), is located in the area offshore of the Naples Bay (Fig. 1).
The sampling site (black dot), Canyon Dohrn and the LTER-MC site in the Gulf of Naples (Central Tyrrhenian Sea, Mediterranean Sea). Created by I. Di Capua with Surfer Ver. 10.7.972 (64-bit); www.goldensoftware.com. Coastline data: NOAA National Geophysical Data Center, Coastline extracted: WLC (World Coast Line), Date Retrieved: 20 March, 2024, http://www.ngdc.noaa.gov/mgg/shorelines/shorelines.html.
After the preprocessing steps, taxonomic filtering (90% threshold) and normalization, 8,562,606 reads representing 8322 ASVs were retained for the COI gene (Supplementary file COI gene). Output of the BLAST searches highlighted that 97% of the sequences (ASVs) found matches with reference sequences with values of similarity of 98–100% that were considered assigned at species level (the full path of taxonomic ranks of reference sequence has been considered valid); 1% of the sequences found matches with reference sequences in the range of 95–97%, and in this case the assignment was considered reliable at the genus level (the path of taxonomic ranks of reference sequence was truncated at genus level); finally, 2% of the sequences found matches with reference sequences with values of similarity lower than 95% (the path of taxonomic ranks was truncated at family or order level). After this step, we applied the ASV clustering (the abundances of ASVs with the same taxonomic path were summed), resulted in a list of 206 taxa (Supplementary file COI).
For the 18S V9, after the preprocessing steps, taxonomic filtering (90% threshold) and normalization, the dataset consisted of 10,045,999 reads, corresponding to 1640 ASVs (Supplementary file 18S V9 gene).The output of the BLAST searches highlighted that 69% of the sequences found a match with reference sequences with values of similarity of 99–100% and were considered assigned at the species level (the full path of taxonomic ranks of reference sequence has been considered valid); about 20% of the sequences found matches with reference sequences in the range of 95–98% and in this case the assignment was considered reliable at genus level (the path of taxonomic ranks was truncated at genus level); 10% of the sequences found matches with reference sequences with values of similarity lower than 95% (the path of taxonomic ranks was truncated at family or order level). After this step, we applied the ASV clustering (the abundances of ASVs with the same taxonomic path were summed), producing a list of 139 taxa (Supplementary file 18S V9).
Using morphological identification, we reported that the zooplankton community constituted of 105 taxa identified at different taxonomic levels (Supplementary file Morpho) (Table 1).
Overall, alpha diversity indexes did not show specific trends. The values, calculated using the post-clustering ASVs tables normalized, highlighted higher species richness values (observed taxa) in the COI gene than in the 18S V9 region and morphological dataset, but quite similar ranges of values were found for the Shannon index in all three datasets (Supplementary Fig. S1a–c).
Taxonomic overview of the three datasets (COI vs V9 vs Morphology)
Overall, in the case of the COI gene 11 holoplanktonic and nine meroplanktonic taxa were found, including ichthyoplankton assemblage (Fig. 2). For the 18S V9 region, eight holoplanktonic and 12 meroplanktonic were found (Table 1). Using morphological identifications, 11 holoplanktonic and 6 meroplanktonic taxa were reported (Table 1). Both molecular and morphological approaches showed that holoplanktonic organisms were more abundant than meroplanktonic ones. Copepoda was the dominant taxon, representing an average of 76% of the total reads in the COI gene, 48% in the18S V9 region, and 78% in microscopy identification. The other crustacean taxa (Amphipoda, Diplostraca, Euphausiacea, Ostracoda) represented an average of 10% in the COI gene, 12% in 18S V9 region, and 14% in microscopy dataset. Lower percentages were found for the holoplankton taxa, including Chaetognatha, Hydromedusae, Pelagiidae, Siphonophorae, Thaliacea, and Appendicularia, were included with lower percentages.
The 18 V9 region generally detected more meroplanktonic larvae than the COI gene dataset. Meroplanktonic larvae (Anthozoa, Bryozoa, Cirripedia, Decapoda, Echinodermata, Hemichordata, Malacostraca, Mollusca, Polychaeta, Others) represented an average of 7% of the total reads in the COI gene and 30% in 18S V9 region. Microscopy counts for the meroplanktonic taxa allowed the identification of fewer taxa at higher taxonomic levels (Mollusca, Polychaeta, Decapoda, and Echinodermata) with a total average abundance of 3% (Table 1). The ichthyoplankton assemblage was mainly assigned to Teleostea (on average, 2% in COI gene and 0.2% in 18S V9 region). In contrast, only a few unidentified fish eggs (2.4 ind. m−3) were counted with microscopy only at LTER-MC (Table 1).
Metazoan distributions across the two stations (Canyon vs LTER-MC)
The distribution of taxa at the two stations showed that in the COI gene, the crustacean dominated all samples, accounting for a total average of 92%. This comprises 93% of samples in Canyon and 90% in LTER-MC. A similar dominance of crustaceans was observed also for the 18S V9 region, with an average value of 79%. Crustacean accounted for 73% in the Canyon, while at LTER-MC it was 85% (Table 1). The second most abundant taxa differed between the molecular markers. In the COI gene dataset, cnidarians (Anthozoa, Hydromedusae, Pelagiidae, and Siphonophorae) account for an average of 3% of reads (Table 1, Fig. 4a). In contrast, in the 18S V9 region dataset, chaetognaths (Sagittoidea) represented the second most abundant phylum with an average of read abundance of 8% (Table 1, Fig. 4b). The analysis of the COI gene and morphology analyses revealed that micro-crustacean copepods were the dominant taxa at both stations (Fig. 3a,c). However, in the 18S V9 region, the dominance of the copepods was most evident in the Canyon. At the same time, at LTER-MC, the group only dominated in the spring sample (MC1_April) with 68.5% (Fig. 3b, Supplementary file 18S V9).
Focusing on non-copepod holoplankton components, it was found that the COI gene and morphological data showed similar distributions. The Ostracoda taxa, mainly belonging to the Order Halocyprida, with Porroecia spinirostris, was more abundant in the Canyon based on COI gene and morphology, but this presence was partially lost in the 18S V9 region (Fig. 4a,b). Similarly, the Diplostraca taxa were more abundant at station LTER-MC based on COI gene and morphology (Fig. 4a,c), but cladocerans (Diplostraca) were absent in the 18S V9 region dataset (Fig. 4b).
Meroplankton was more abundant at the coastal station LTER-MC than at Canyon on the COI gene, with the Cirripedia and Decapoda as the main taxa (Table 1). At the Canyon, meroplankton was mainly represented by Echinodermata (Echinoidea, Ophiuroidea, Holothuroidea) and Polychaeta, with the presence of the species Magelona lusitanica being detected in the layer 0–200 m in spring using the COI gene.
The two genetic markers showed different accuracies in identifying fish at the species level. The 18S V9 region had lower resolution power compared to the COI gene. Using the COI gene, we were able to identify several fish species at LTER-MC, including Mullus barbatus, Engraulis encrasicolus, Trachurus mediterraneus, Apogon imberbis, Serranus hepatus, Anguilla rostrata, Auxis rochei, Megalops atlanticus, and Sarpa salpa. Additionally, at the Canyon, we found Boops boops in the layer 0–50 m and the deep-sea fish Ceratoscopelus maderensis in the layer 0–200 m.
Some taxonomic groups were detected using only one of the two markers: the 18 V9 region allowed for the detection of Appendicularia (Fig. 4b), but this was lost with the COI gene (Fig. 4a). Within appendicularians, we reported Oikopleura dioica and Oikopleura sp., as well as the Thaliacea, which include Doliolum denticulatum and D. nationalis (Supplementary file 18S V9). On the other hand, the 18 V9 region failed to detect the Amphipoda, Diplostraca, Pelagiidae, and Siphonophorae (Fig. 4b,a, Table 1).
Overall, the COI dataset shared a core of 23 species, whereas only 10 were shared among the 18S V9 region samples. However, both datasets highlighted that each sample showed a low number of exclusive taxa except for the sample C1_200 at Canyon, which had 39 exclusive species with the COI region and 14 with V9 (Supplementary Fig. S2a–c).
Focus on dominant taxa (COI vs morphology)
Similar results were observed for the identification of Copepoda using the COI gene and morphology-based identification, with respectively 63 species and 72 species identified. Thirty four copepod species from 14 families were identified at both stations, LTER-MC and Canyon, using both approaches, with 31 being calanoids (Fig. 5a,b, Supplementary Table S1). The distribution details of these 34 species showed recurrent presence in the COI data set (Fig. 5a, Supplementary Table S1).
The dominance of the calanoids Acartia clausi and Temora stylifera was confirmed by both the COI gene and morphological approaches. Moreover, other calanoids, eight species of Clausocalanus and four of Calocalanus were identified (Fig. 5a,b). As expected, the COI gene distinguished the three calanoid species Paracalanus quasimodo, P. parvus, and P. indicus. In contrast, the microscopic approach identified them as a unique species complex (P. parvus complex), with a total abundance of 1273 ind. m−3. The non-indigenous calanoid species Pseudodiaptomus marinus was detected using the COI gene only at LTER-MC in the June sample (Supplementary Table S1). Additionally, several meso-bathypelagic calanoid species (Haloptilus longicoris, Heterorhabdus papilliger, Lucicutia flavicornis, Pleuromamma abdominalis, Neocalanus gracilis) were exclusively detected and identified at the Canyon (Fig. 5a,b).
The COI gene and morphology showed different results for cyclopoid copepod species, specifically within the Oithona and Coryaceus genera (Supplementary file COI and 18S V9). The COI gene indicated only the presence of O. davisae, while morphological identification revealed the presence of eight different species, excluding O. davisae (Supplementary File Morpho) and any corycaeids species were detected. Both approaches detected Oncaea (Fig. 5a,b) and Triconia at species level (Supplementary Table S1).
The larvae of Euphausiacea, including calyptopis and eggs, were reported in morphological dataset with a very low abundance (1.1 ind. m−3), but the COI gene successfully identified 11 Euphasiacea species across five genera. Specifically, at the Canyon, nine species from four genera were detected, with five of them exclusively found at the deeper layer of the Canyon (Meganyctiphanes norvegica, Nematoscelis atlantica, N. megalops, N. difficilis, Thysanoessa raschii) (Fig. 6a). Euphausia krohni, which was abundant in all Canyon samples, was only present in two samples at the LTER-MC station with very low abundance. Nycthiphanes simplex, another commonly represented euphausiid, showed an opposite distribution pattern, dominating at the LTER-MC rather than the Canyon samples (Fig. 6a).
Finally, the morphological approach to identifying cnidarians was ineffective, but only the COI gene revealed the presence of 19 gelatinous taxa (Fig. 6b). These included Hydromedusae, six species of Siphonophorae, and one species of Pelagidae (Pelagia noctiluca) (Fig. 6b).
Discussion
Understanding the dynamics of the zooplankton community in the Mediterranean basin is essential to properly address local anthropogenic pressures and global climate change challenges on marine ecosystems. At the LTER-MC, morphological data of phytoplankton and zooplankton components have been collected weekly since 1984, also linking them with oceanographic, chemical, and physical parameters29. However, the standard procedures for monitoring marine communities in time series monitoring and field research must integrate advanced molecular approaches (DNA barcoding and metabarcoding) to expand knowledge and expedite routine actions and studies30. The implementation of environmental DNA metabarcoding requires understanding its limits and potentialities in terms of ecological inferences, as well as harmonizing protocols with the establishment of best practices to apply to different marine ecosystem. This study is the second using eDNA to analyze metazoan communities from the Gulf of Naples11, and are currently undergoing various studies to enhance and supplement traditional monitoring methods with DNA-based approaches such as metabarcoding, metagenomics, and metatranscriptomics in the framework of the Naples Ecological Research augmented observatory Project (NEREA) (https://www.nerea-observatory.org/nereasampling).
COI gene vs 18S V9 region
The combined use of the molecular markers COI and 18S is a common practice in marine zooplankton metabarcoding-based assessments10. The COI gene, in particular, is known to reveal a greater number of metazoan species and provide better taxonomic resolution compared to the 18S regions, which have highly conserved primer sites, allowing for amplification of a wide range of metazoan and protists17,31. Our results further demonstrate the value of this approach, showing that using both markers identifies more metazoan zooplanktonic taxa than using a single molecular marker. This leads to higher taxonomic resolution and better assignment quality, enhancing the accuracy and reliability of our findings.
The COI gene detected more taxa (206) than the 18S V9 region (139). It also provided more accurate assignments, with 97% of COI sequences being assigned at the species level, compared to 69% for the 18S V9 region. At a higher taxonomic level, the two markers performed similarly, with the main differences being the detection of Diplostraca and Siphonophorae only with the COI gene and Appendicularia only with the 18S V9 region. Difficulties detecting Diplostraca with an 18S marker have been reported previously, including when using the 18S V4 region10,11. Unlike what we observed in the 18S V9 region, Siphonophorae can also be detected in the 18S V4 region11. The absence of Appendicularia in the taxonomic profile from the COI gene was expected, as reference COI barcodes exist for only 8% of their described diversity32, and PCR primers for the COI barcodes have been reported to fail in several cases32,33.
Our study confirmed that using two molecular markers in organismal eDNA assessments improves species detection. This helps to reduce biases related to different PCR primer amplification affinities among organisms and the varying taxonomic coverage of reference sequences in databases. However, our results also suggest that caution is needed when choosing primer pairs. Universal primers that amplify alternative regions within the same molecular marker, such as the V4 and V9 regions in the 18S marker, can detect different phyla.
However, the success of taxonomic assignments in eDNA assessments depends on the quality and quantity of reference barcodes in the databases. Several initiatives are being undertaken to create a comprehensive and curated COI reference database for marine zooplankton32. On the other hand, the resources available for 18S rRNA mainly focus on protists34 or are less curated35. In our case, having COI barcode references previously generated for zooplankton specimens collected in the studied region36,37, and reference therein] could have improved the taxonomic assignments of COI gene at the species level compared to the 18S V9 region. This would also have provided a taxonomic profile more consistent with the morphological observations.
Organismal eDNA vs morphology
Our results show that organismal eDNA data closely matches the findings from morphological identification, confirming previous studies of zooplankton communities at different spatial and temporal scales32,33,38. Overall, there is strong agreement between morphological and molecular results for holoplanktonic organisms. However, morphology-based results are slightly more accurate, especially for the important and abundant group of micro-crustacean copepods, which reported a higher total number of species (72). On the other hand, the COI gene consistently shows the presence of copepods in all samples, likely due to the inclusion of nauplii and/or copepodid stages in the molecular results, which were not identified and/or were lost using the traditional morphological approach. Out of 34 species shared between the molecular identification with the COI gene and the morphological list, 31 were belonged to the calanoid order (Supplementary Material, Table S1). The morphological list includes only nine species not detected by the COI gene. Most of these species belong to the cyclopoid order of the Oithona and Corycaeus genera, that do not yet have available barcodes. Despite this, the COI gene provides correct identification in the case of the cryptic species Paracalanus quasimodo, P. parvus, and P. indicus37.
The combination of molecular and morphological approaches improves the understanding gained solely from morphological studies, especially when studying the meroplanktonic component. It was expected that eDNA would outperform morphology-based surveys for screening meroplankton, as demonstrated in previous studies39,40. Molecular identification is particularly useful for identifying larval stages of benthic species, which can be challenging or impossible using morphology-based methods. Previous knowledge of the diversity of meroplanktonic metazoans in the Gulf of Naples study areas was limited, mainly including morphological identification of decapod larvae in spring and larvae of cirripedes and echinoderms in winter37. However, this study revealed a greater diversity, confirming and expanding our previous eDNA results obtained using the 18S V4 region11, with overall the presence of 13 phyla.
Biodiversity and ecological patterns
Our study provides for the first overview of zooplankton metazoan communities and diversity in the Canyon Dohrn site in two different depth layers. With a length of approximately 25 km and a width of nearly 2 km, the Canyon Dohrn represents a crucial bathymetric feature of the Gulf of Naples41,42. The head of the Canyon is steep, and gets very close to the coastline, consequently supporting the exchanges between the deep sea and the shelf area35,43, and reference therein]. A recent study investigation, performed by visual identification of ichthyoplankton, reported the presence of larvae belonging to 11 families and of eggs belonging to five families44. In addition, the deep-sea benthic community was also explored based on an ROV survey by45 et al., highlighting the Dohrn Canyon’s high biodiversity and evidence of severe anthropogenic impacts. The authors also reported the co-existence of living specimens of the bivalves with the cold-water corals, which has never been previously reported in the Mediterranean Basin. In contrast, no planktonic surveys exist, but our preliminary findings revealed differences between the two sampling sites (Canyon and LTER-MC), highlighting in the submarine Canyon Dohrn the presence of deeper-living zooplankton species such as meso-bathypelagic copepod species, euphausiids, and hydrozoan-groups. These species, often challenging to identify or poorly characterized through morphological methods, were successfully identified at the species level using the COI gene46. Nevertheless, euphausiids are hard to identify using only morphology beyond the genus level, particularly at larval stages36. Of note, among the euphasiids, we recently obtained the barcodes for the Euphausia krohnii, Nematoscelis megalops, and the genus Nyctiphanes in this study area36.
Morphological features are often insufficient for accurate identification beyond the family level. It is difficult, and therefore impossible, to morphologically distinguish the larval stages of many benthic species and within groups of gelatinous holoplankton, especially when using samples fixed with ethanol (Di Capua, personal communication). In these cases, the use of eDNA organismal can help to improve our understanding of the presence and distribution of these taxa, especially in coastal areas, such as the LTER-MC site, and also better to understand the relationship between benthic and pelagic coupling.
Molecular data, however, have revealed the presence of 20 cnidarian species belonging to Anthozoa, Hydromedusae, Siphonophorae, and Pelagiidae. Notably, COI gene analysis has detected medusae P. noctiluca in the 200–0 m of the Canyon in April and at LTER-MC in May, where they are expected to feed on spring zooplankton and ichthyoplankton47. These findings confirm the significant role of submarine canyons in regulating vertical matter fluxes and facilitating exchanges between deeper layers and the surface44. The connection between the offshore submarine Canyon station and the coastal station LTER-MC depends on specific surface and deep circulation patterns, which may vary depending on the seasonal forcing scheme48. Furthermore, the P. noctiluca ephyrae in both sites aligns with the hypothesis of seasonal vertical migration of this species in the Mediterranean Sea49.
Overall, the fish in the area belong to 14 species across nine families, including commercially important ones like Engraulis encrasicolus and Mullus barbatus. Recently, five of these families (Engraulidae, Gobiidae, Myctophidae, Serranidae, Carangidae) have been identified in the sampling area through morphological-based identification44.
The calanoid copepods, such as Acartia clausi and Temora stylifera have been confirmed as the most abundant species using COI gene and morphology, confirming the ecological significance of these two species in this neritic ecosystem48,49,52. Additionally, our data showed a recurrent copepod community structure in both sites in the depth layer 50–0 m, probably related to the water mixing process in the surface mixed layer at 30–50 m depth44,48.
Surveys of non-indigenous species (NIS) are especially important in LTER sites, where knowledge of species composition can aid in the early detection of NIS and their invasive potential. Organismal eDNA has proven effective in tracing rare species and larval stages, with several studies demonstrating its accuracy in detecting NIS in marine environments11,53,54. In our survey, the COI gene revealed 12 new records of metazoans in the Mediterranean Sea, including holoplankton taxa such as Larsonia pterophylla, Euchaeta rimana, Oithona davisae, Nyctiphanes australis, Nyctiphanes simplex, Proceroecia procera, and meroplankton taxa including Magelona lusitanica, Balanus trigonus, Psammechinus miliaris, Glossobalanus marginatus, Aplysia Juliana, and Philine indistincta. Identifying these previously undocumented species needs to be confirmed in future assessments, but raises concerns about the impact of climate change and other anthropogenic pressures on biodiversity in the Mediterranean Sea. Notably, the polychaete M. lusitanica has already been molecularly characterized in nearby Atlantic waters47, suggesting it may be a new record for the Mediterranean Sea, as recently observed for other non-indigenous polychetes along the coasts of Italy55. Additionally, our results confirmed the presence of the non-indigenous copepod Pseudodiaptomus marinus at the long-term coastal station56, and the COI gene detected a new non-indigenous species, Oithona davisae for the first time.
Our research confirms that using organismal environmental DNA (eDNA) is an effective molecular approach for studying the diversity and distribution of metazoan communities, and it is an improvement over traditional morphology-based approaches. Using a multi-loci strategy, we could cover a wide range of metazoan phyla. However, the choice of primer pairs can significantly impact the taxonomic results. We found that the COI gene identified more holo- and meroplanktonic organisms than the 18S V9 region and provided more accurate classifications in according to morphological identification. Specifically, 97% of COI sequences were matched to specific species, compared to 69% for the 18S V9 region. Both markers performed similarly when identifying groups of organisms at high taxonomic levels (es. subclass, order), but they complemented each other. Despite some limitations, organismal eDNA data provided valuable insights into hidden biodiversity, especially for the gelatinous and meroplankton components, including juvenile or larval stages that cannot be identified through morphology-based approaches. Additionally, the use of local barcode references obtained from COI analysis of zooplankton specimens probably contributed to the accuracy of this marker in producing a taxonomic profile that closely matches morphological observations. This highlights the importance of enhancing the reference databases globally and locally, emphasizing their crucial role in ensuring reliable ecological inferences derived from metabarcoding data across various sample types. Furthermore, it identified signs of potential ecosystem deterioration by detecting new entries and non-indigenous species (NIS). Overall, applying the organismal eDNA approach at LTER sites increases its potential impact, as it quickly identifies changes in zooplanktonic community biodiversity and guides the optimization of biomonitoring approaches.
Materials and methods
Study area
The site LTER-MC (40° 48.5′ N; Long. 14° 15′ E) is a coastal station (depth ca. 75 m), located two nautical miles from the city of Naples, is influenced by the eutrophic coastal zone and also the oligotrophic waters of the mid Tyrrhenian Sea40. LTER-MC has long been sampled for plankton and other parameters since 1984, and the dataset represents one of the few plankton time series available in the Mediterranean Sea (https://www.st.nmfs.noaa.gov/copepod/time-series/it-30101/). All these observations highlight a strong seasonal signature of the biodiversity pattern of the area, and zooplankton communities are diversified and stable throughout the year52. The Canyon site (C) (40° 36.232′ N; 14° 08.012′ E) located approximately 12 nautical miles offshore from the Naples metropolitan area, was originated by ancient eruptive events involving the area offshore the Naples Bay57. From a geological point of view, the Canyon was explored, integrating stratigraphic study of geophysical and geological to provide information for evaluating and mitigating natural hazards (i.e., volcanism, seafloor stability) in managing densely populated coastal areas57. Biodiversity and ecology knowledge of Canyons are limited and focused on deep-sea benthic communities. Nevertheless, based on the ROV survey, Taviani et al. (2019) highlighted that, despite evidence of severe anthropogenic impacts, the Dohrn Canyon was a highly diverse habitat, also reporting the co-existence of living specimens of the bivalves with the cold-water corals, which has never been previously reported in the Mediterranean Basin.
Sampling strategy and sample treatment
Sampling of communities was performed during 2019: four for the Canyon station (C1_50; C1_200; C2_50; C2_200) and four for the LTER-MC station (MC1_50; MC2_50; MC3_50; MC4_50) (Fig. 1). The sampling covered three different seasons; in particular, the LTER-MC station covered three different seasons (autumn, spring, winter), while two seasons (autumn, spring) for the Canyon (C) station. Details regarding station label and surface environmental data are reported in Supplementary File final, Supplementary File sampling site. Zooplankton samples were collected with a double WP2 plankton net with a mouth area of 0.25 m2 and a mesh aperture of 200 μm, each equipped with a 500 mL plexiglass filtering cod-end. This double WP2 plankton net allowed two simultaneous samples for each tow for morphology-based and molecular analyses. The net was towed vertically from 50 m to the surface at LTER-MC, from 200 m to the surface, and from 50 to the surface at Canyon. The filtered water volume was calculated using a mechanical flowmeter (Hydrobios). Upon collection, zooplankton samples were immediately preserved in 95% ethanol for taxonomic identification. For metabarcoding, samples were prefiltered on a 2000 μm filter mesh to remove larger organisms, then the samples were filtered on a 200-µm filter mesh; the resulting pellets of bulk zooplankton were collected and placed into 2 mL cryogenic vials, immediately flash-frozen in liquid nitrogen and then stored at − 80 °C until later DNA extraction.
Morphology-based approach
In the laboratory, the ethanol-fixed samples were concentrated and re-suspended in ethanol 200 mL volume. Aliquots ranging from 1/4 to 1/32 according to sample density, were taken initially by using the Huntsman beaker subsampling technique and successively using a large bore graduated pipette after careful mixing43. These aliquots were transferred in a mini-Bogorov chamber (10 mL) and examined under a stereomicroscope (Leica M165C). For all copepods, adult females and males were separately identified at species level. Depending on the species, copepodites (CII-CV) were identified at the species level in most cases or grouped at genus or family level (e.g. Calocalanus, Oithona, Oncaeidae, Corycaeidae). Copepod species abundances (ind. m−3) reported here correspond to the whole population (adult females and males, and copepodites), unless otherwise indicated. Among other zooplankters, some groups were identified at the species level (e.g. cladocerans, chaetognaths, siphonophors), while other holoplankton (e.g. amphipods, mysids, ostracods, euphasids, pteropods, hydromedusea) and meroplankton taxa (decapods, cirripedes, echinoderms, bivalvs, molluscs, gastropods) were identified at higher taxonomic levels. All zooplankters were identified and counted, then the rest of the sample was checked for the presence of rare species. The calculation of abundance (ind. m−3) needs to consider the number of individuals counted in the subsample (n), which fraction of the original sample does the counted sub-sample represent (k), and the water volume filtered by the sampling net (V, m3), according to the formula: ind. m−3 = (n × k)/V m3.
Molecular approach
All samples were shipped on dry ice to Genoscope Sequencing Laboratory (France) where DNA extractions and sequencing were performed. DNA extraction from each zooplankton pellet was performed using Cryogrinding of two different pellets separately, followed by Nucleospin RNA/DNA co-extraction (Macherey Nagel). For each sample, PCR amplifications were performed in triplicates (5–10 ng DNA per amplification) that were then merged. Two different metabarcoding markers were amplified: mitochondrial cytochrome c oxidase gene (COI) and the V9 region of the nuclear 18S rRNA (V9). For COI gene we amplified a fragment of ca. 313 bp using a primer pair described in Leray et al. (2013) (forward m1COIint 5′-GGWACWGGWTGAACWGTWTAYCCYCC-3′ and reverse jgHCO2198 5′-TAIACYTCIGGRTGICCRAARAAYCA-3′. For V9 region of 18S rRNA were used a primer pair described in31 (forward 1389F 5′-TTGTACACACCGCCC1510R-3 and reverse 1510R 5′-CCTTCYGCAGGTTCACCTAC-3′) amplifying a ca. 136 bp fragment. Illumina HiSeq sequencing paired-end were performed by Genoscope as described in58 using (2 × 250 bp) for the COI gene and (2 × 150 bp) for the V9 18S region.
Data preprocessing and taxonomic assignment
Illumina paired-end raw reads were processed to generate ASVs using the DADA2 R package59, with some modifications for the COI gene and the V9 18S markers.
COI: Quality check was performed with the vsearch –fastq_stats command (Rognes et al. 2016). Primers were trimmed with cutadapt60. After primer removal, forward and reverse reads were truncated based on the Quality score plots (forward: 210 nucleotides; reverse: 200 bp respectively) and then used to train the error model using machine learning approach. Forward and reverse reads were then separately reduced in sequence variants using error rates calculated in the previous step for forward and reverse applying pseudo pooling option. Finally, forward, and reverse reads were merged to obtain the full denoised sequences. The ASV table was further filtered based on expected amplicons length (309–317 bp) and checked for presence of chimera (option method = consensus in the removeBimeraDenovo function in DADA2). ASVs were taxonomically assigned using standalone Blast 2.8.1 in the blast + suite61,62 against a custom database integrating MetaCOXI and MetaZooGene reference database63,64. In addition, we added a selection of 13,507 protist and macroalgae sequences (organisms not included in MetaCOXI and MetaZooGene) obtained from GenBank (full list is available in the Supplementary File final) and 23 copepod sequences from the Gulf of Naples (GenBank accessions MZ710171 to MZ710194). Assignments with similarity lower than 90% and query coverage lower than 250 bp were excluded from Blast results. Finally, ASVs assigned to non-marine taxa (Arachnidae, Collembola, Insecta, Amphibia, Craniata and terrestrial Mammalia and protists) were removed from the analyses.
18 V9: Quality check was performed with the vsearch –fastq_stats command60. Primers were trimmed with cutadapt65. After primer removal, forward and reverse reads were truncated based on the Quality score plots (forward: 120 nucleotides; reverse: 130 bp respectively). Denoising steps were performed with the same procedure described for the COI gene. The ASV table was filtered based on expected amplicons length (125–150 bp) and checked for presence of chimera as performed for COI gene. Taxonomic assignment of ASVs was performed using standalone Blast 2.8.1 in the blast + suite61,62 against a custom version of PR2_4.14.3 reference Database34. This Database was integrated with 330 sequences of Protists from the Gulf of Naples (Supplementary File PR2) and 30 other environmental marine Fungi sequences66. Assignments with lower than 90% similarity and query coverage lower than 90 bp were excluded from Blast results. ASVs assigned to non-marine taxa (Arachnidae, Collembola, Insecta, Amphibia, Craniata and terrestrial Mammalia and protists) were removed from the analyses.
Post ASV clustering
In metazoan diversity assessments, especially in comparison with counts from morphological data, the assumption that each ASV is a biological entity (species) could produce a strong overestimation. Nevertheless, biological variations (intra genomic, intra individual, intra species variations) remain in the ASVs table. After the taxonomic assignment with BLAST, we applied a taxonomy-guided clustering of ASVs that share the same taxonomic path to overcome this pattern. We first ranked the assignments of ASVs into three levels based on the similarity with the reference from the BLAST output.
In the case of COI gene: (i) assignments showing similarity with the reference sequence in the range 98–100% were considered assigned at species level (meaning that the full path of taxonomic ranks of reference sequence has been considered valid); (ii) assignments showing similarity with the reference sequence in the range 95–97% were considered reliable at genus level (meaning that the path of taxonomy of reference sequence was truncated at genus level); (iii) assignments showing similarity with the reference sequences lower than 95% were considered reliable at higher levels (the path of taxonomy was truncated at family or order level). After that, the ASVs with the same taxonomic path were collapsed together. In the case of several ASVs with taxonomic path truncated at the same genus (or family/order), they have been collapsed together.
The same approach but different thresholds were applied for the 18S V9 region: (i) assignments showing similarity with the reference sequence in the range 99–100% were considered assigned at species level (meaning that the full path of taxonomic ranks has been considered valid); (ii) assignments showing similarity with the reference sequence in the range 95–98% were considered reliable at genus level (meaning that the path of taxonomy was truncated at genus level); (iii) assignments showing similarity with the reference sequences lower than 95% were considered reliable at higher levels (the path of taxonomy was truncated at higher level). After that, the ASVs with the same taxonomic path were collapsed together. In the case of several ASVs with taxonomic path truncated at the same genus (or family/order), they have been collapsed together.
Data analysis
Normalization of the datasets was applied to counterbalance the potential biases in the analyses related to the different number of reads obtained in the sequencing of the samples. Thus, after the post ASVs clustering step, the COI gene and 18S V9 tables were subsampled to the lowest number of reads (1,070,326 for the COI and 58,252 for the 18S V9) using the function rrarefy in vegan R package67. The phyloseq68 and ggplot269 R packages were used to generate alpha diversity descriptors (Richness (number of observed taxa) and Shannon entropy index) and the plots of the results. After logarithmic transformation, cluster analyses of molecular and morphological datasets were performed based on a Bray–Curtis dissimilarity matrix with vegan package. Part of the analyses (the comparison with morphological data and the generation of a phylogenetic tree) were performed only using the COI dataset representing the most frequently used and powerful barcode region, also providing a large number of curated reference sequences for a successful species-level identification of marine zooplankton32. The phylogenetic tree was built (only for COI dataset) using a ASV representative sequence and two protist sequences detected in our dataset as outgroups (Hematodinium sp. and Azadinium dalianens). The alignment was performed with online MAFFT version 769 (https://mafft.cbrc.jp/alignment/server/index.html) then visualized in SeaView70. The maximum-likelihood tree was built in FastTree71 with generalized time-reversible models and visualized in iTOL (https://itol.embl.de/) 72. In addition, the molecular and morphological taxa lists were annotated as holo or meroplankton.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Files (Sampling sites, COI gene, 18 S V9 region, Morpho). The metabarcoding data generated in this study have been deposited in the ENA (European Nucleotide Archive, http://www.ebi.ac.uk/ena) under the umbrella project permanent identifier PRJEB74649 ‘The NEREA Augmented Observatory: a holistic approach to ecological long-term research’.
References
Gomes, L. F. et al. Zooplankton functional-approach studies in continental aquatic environments: A systematic review. Aquat. Ecol. 53, 191–203 (2019).
Lomartire, S., Marques, J. C. & Gonçalves, A. M. The key role of zooplankton in ecosystem services: A perspective of interaction between zooplankton and fish recruitment. Ecol. Ind. 129, 107867 (2021).
Hobday, A. J., Okey, T. A., Poloczanska, E. S. et al. Impacts of climate change on Australian marine life. Report to the Australian Greenhouse Office, Canberra (2006).
Carter, J. L., Schindler, D. E. & Francis, T. B. Effects of climate change on zooplankton community interactions in an Alaskan lake. Clim Change Responses 4, 1–12 (2017).
Ratnarajah, L. et al. Monitoring and modelling marine zooplankton in a changing climate. Nat. Commun. 14(1), 564 (2023).
Couret, M., Landeira, J. M., del Pino, Á. S. & Hernández-León, S. A 50-year (1971–2021) mesozooplankton biomass data collection in the Canary Current System: Base line, gaps, trends, and future prospect. Prog. Oceanogr. 216, 103073 (2023).
Harvey, C., Garfield, N. T., Williams, G., et al. Ecosystem status report of the California current for 2019–20: A summary of ecosystem indicators compiled by the California current integrated ecosystem assessment team (CCIEA) (2020).
Phillips, E. M. et al. Spatiotemporal variability of Euphausiids in the California Current Ecosystem: Insights from a recently developed time series. ICES J. Mar. Sci. 79(4), 1312–1326 (2022).
Haubrock, P. J. et al. Long-term trends in abundances of non-native species across biomes, realms, and taxonomic groups in Europe. Sci. Total Environ. 884, 163808 (2023).
Zhao, L., Zhang, X. & Huang, Y. DNA metabarcoding of zooplankton communities: Species diversity and seasonal variation revealed by 18S rRNA and COI. PeerJ 19, e11057 (2021).
Di Capua, I., Piredda, R. & Zingone, A. Metazoan diversity and seasonality through eDNA metabarcoding at a Mediterranean long-term ecological research site. ICES J. Mar. Sci. 78, 3303–3316 (2021).
Cristescu, M. E. From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 29(10), 566–571 (2014).
Leray, M. & Knowlton, N. Visualizing patterns of marine eukaryotic diversity from metabarcoding data using QIIME. Methods Mol. Biol. 35, 219–235 (2016).
Schenk, J. & Fontaneto, D. Biodiversity analyses in freshwater meiofauna through DNA sequence data. Hydrobiologia 847, 2597–2611 (2020).
Song, C., Choi, H. & Jeon, M. S. Zooplankton diversity monitoring strategy for the urban coastal region using metabarcoding analysis. Sci. Rep. 11, 24339 (2021).
Qihang, L. et al. Metabarcoding survey of meroplankton communities in the South China Sea and Philippine Sea: Shedding light on inter-basin biogeography in the West Pacific. Front. Mar. Sci. 9, 968666 (2022).
Bucklin, A., Batta-Lona, P. & O’Brien, T. D. COI metabarcoding of zooplankton species diversity for time-series monitoring of the NW Atlantic continental shelf. Front. Mar. Sci. 9, 867893 (2022).
Feng, Y. et al. COI metabarcoding better reveals the seasonal variations in the zooplankton community in the western Pacific Warm Pool. Ecol. Indic. 156, 111183 (2023).
Moutinho, J. et al. Assessing the seasonal and spatial dynamics of zooplankton through DNA metabarcoding in a temperate estuary. Animals 13(24), 3876 (2023).
Berry, T. E., Saunders, B. J. & Bunce, M. Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet. 15(2), e1007943 (2019).
Westfall, K. M., Therriault, T. W. & Abbott, C. L. A new approach to molecular biosurveillance of invasive species using DNA metabarcoding. Glob. Change Biol. 26(2), 1012–1022 (2020).
Duarte, S., Vieira, P. E., Lavrador, A. S. & Costa, F. O. Status and prospects of marine NIS detection and monitoring through (e) DNA metabarcoding. Sci. Total Environ. 751, 141729 (2021).
Goldberg, C. S. et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evolut. 7(11), 1299–1307 (2016).
Van der Heyde, M., Bunce, M. & Nevill, P. Key factors to consider in the use of environmental DNA metabarcoding to monitor terrestrial ecological restoration. Sci. Total Environ. 848, 157617 (2022).
Grey, E. K. et al. Effects of sampling effort on biodiversity patterns estimated from environmental DNA metabarcoding surveys. Sci. Rep. 8(1), 8843 (2018).
Coll, M. et al. The biodiversity of the Mediterranean Sea: Estimates, patterns, and threats. PLoS ONE 5(8), e11842 (2010).
Lionello, P. & Scarascia, L. The relation between climate change in the Mediterranean region and global warming. Reg. Environ. Change 18, 1481–1493 (2018).
Cramer, W. et al. Climate change and interconnected risks to sustainable development in the Mediterranean. Nat. Clim. Change 8(11), 972–980 (2018).
Zingone, A. et al. Time series and beyond: Multifaceted plankton research at a marine Mediterranean LTER site. Nat. Conserv. 34, 273–310 (2019).
Takahashi, M. et al. Aquatic environmental DNA: A review of the macro-organismal biomonitoring revolution. Sci. Total Environ. 873, 162322 (2023).
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W. & Huse, S. M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4(7), e6372 (2009).
Bucklin, A. et al. Toward a global reference database of COI barcodes for marine zooplankton. Mar. Biol. 168(6), 78 (2021).
Schroeder, A. et al. Suitability of a dual COI marker for marine zooplankton DNA metabarcoding. Mar. Environ. Res. 170, 105444 (2021).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41(D1), D597–D604 (2012).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41(D1), D590–D596 (2012).
Di Capua, I., Piredda, R. & Uttieri, M. Is integrated taxonomy useful to study diversity and ecology? An example from crustacean zooplankton at the Long-Term ecological research site MareChiara (LTER-MC). Mar. Ecol. 44, e12752 (2023).
Di Capua, I., D’Angiolo, R. & Carotenuto, I. From phenotypes to genotypes and back: Toward an integrated evaluation of biodiversity in calanoid copepods. Front. Mar. Sci. 9, 833089 (2022).
Bucklin, A., Yeh, H. D. & Wiebe, P. H. Time-series metabarcoding analysis of zooplankton diversity of the NW Atlantic continental shelf. ICES J. Mar. Sci. 76(4), 1162 (2019).
Ershova, E. A. et al. Metabarcoding as a quantitative tool for estimating biodiversity and relative biomass of marine zooplankton. ICES J. Mar. Sci. 78(9), 3342–3355 (2021).
Moutinho, J., Carreira-Flores, D., Gomes, P. T., Costa, F. O. & Duarte, S. Assessing the seasonal and spatial dynamics of zooplankton through DNA metabarcoding in a temperate estuary. Animals 13(24), 3876 (2023).
Aiello, G., Budillon, F. & Tonielli, R. Marine geology and morphobathymetry in the Bay of Naples (South-Eastern Tyrrhenian Sea, Italy). In Mediterranean Ecosystems—Structures and Processes (eds Faranda, F. M. et al.) 1–8 (Springer, 2001).
Cianelli, D., Uttieri, M. & Zambianchi, E. Dynamics of a very special Mediterranean coastal area: The Gulf of Naples. In Mediterranean Ecosystems: Dynamics: Management and Conservation (ed. Williams, G. S.) 129–150 (Nova Science Publishers, Inc., 2012).
Canals, M. et al. Integrated study of Mediterranean deep sea canyons: Novel results and future challenges. Prog. Oceanogr. 118, 1–27 (2013).
Ciannelli, L., Cannavacciuolo, A. & Cianelli, D. Ichthyoplankton assemblages and physical characteristics of two submarine canyons in the south central Tyrrhenian Sea. Fish. Oceanogr. 31, 480–549 (2022).
Taviani, M. et al. A unique and threatened deep water coral-bivalve biotope new to the Mediterranean Sea offshore the Naples megalopolis. Sci. Rep. 9(1), 3411 (2019).
Holst, S., Heins, A. & Laakmann, S. Morphological and molecular diagnostic species characters of Staurozoa (Cnidaria) collected on the coast of Helgoland (German Bight, North Sea). Mar. Biodivers. 49, 1775–1797 (2019).
Canepa, A. et al. Pelagia noctiluca in the Mediterranean Sea. In Jellyfish Bloom (eds Pitt, K. A. & Lucas, C. H.). 237–266. https://doi.org/10.1007/978-94-007-7015-7_11 (Springer, Netherlands, 2014).
Cianelli, D. et al. Inshore/offshore water exchange in the Gulf of Naples. J. Mar. Syst. 145, 37–52 (2015).
Canepa, A., Fuentes, V., Sabatés, A., Piraino, S., Boero, F. & Gili, J.M. (2014) Pelagia noctiluca in the Mediterranean Sea. In: Pitt, K.A. & Lucas, C.H. (Eds.), Jellyfish Bloom. Springer, Netherlands, pp. 237–266.
Di Capua, I. & Mazzocchi, M. G. Non-predatory mortality in Mediterranean coastal copepods. Mar. Biol. 164, 198 (2017).
Di Capua, I. & Mazzocchi, M. G. Population structure of Centropages typicus and Temora stylifera, in relation to different environmental conditions. ICES J. Mar. Sci. 61, 632–644 (2004).
Mazzocchi, M. G. et al. Coastal mesozooplankton respond to decadal environmental changes via community restructuring. Mar. Ecol. 44, e012746 (2023).
Aylagas, E., Borja, Á., Irigoien, X. & Rodríguez-Ezpeleta, N. Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment. Front. Mar. Sci. 3, 96 (2016).
Couton, M. et al. Metabarcoding on planktonic larval stages: An efficient approach for detecting and investigating life cycle dynamics of benthic aliens. Manag. Biol. Invasions 10(4), 657–689 (2019).
Langeneck, J. et al. Non-indigenous polychaetes along the coasts of Italy: A critical review. Mediterr. Mar. Sci. 21(2), 238–275 (2020).
Uttieri, M., Aguzzi, L. & Zervoudaki, S. WGEUROBUS–Working Group. Towards a EURopean OBservatory of the non-indigenous calanoid copepod Pseudodiaptomus marinUS. Biol. Invasions 22, 1–22 (2020).
Aiello, G., Iorio, M., Molisso, F. & Sacchi, M. Integrated morpho-bathymetric, seismic-stratigraphic, and sedimentological data on the Dohrn Canyon (Naples Bay, Southern Tyrrhenian Sea): Relationships with volcanism and tectonics. Geosciences 10(8), 319 (2020).
Alberti, A. et al. Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data 4(1), 1–20 (2017).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Rognes, T. et al. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 (1990).
Camacho, C. BLAST plus: architecture and applications. BMC Bioinf. 10(421), 1 (2009).
Balech, B. et al. MetaCOXI: An integrated collection of metazoan mitochondrial cytochrome oxidase subunit-I DNA sequences. Database 2022, baab084 (2022).
Bucklin, A., Peijnenburg, K. T. C. A. & Weydmann-Zwolicka, A. Toward a global reference database of COI barcodes for marine zooplankton. Mar. Biol. 168, 78 (2021).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17(1), 10–12 (2011).
Richards, T. A., Leonard, G. & Chambouvet, A. Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc. R. Soc. B Biol. Sci. 282, 20152243 (2015).
Oksanen, J., Blanchet, F. & O’Hara, R. B. Vegan: Community Ecology Package (2013).
McMurdie, P. J. & Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evolut. 30, 772–780 (2013).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evolut. 27, 221–224 (2010).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, 256–259 (2019).
Acknowledgements
We thank all LTER-MC and NEREA teams, and the crew of the R/V Vettoria for assistance during the work at sea. The Long-Term Ecological Research site MareChiara program is funded by Stazione Zoologica Anton Dohrn, Naples (Italy). The NEREA activities have received funding from the European Union’s Horizon 2020 600 research and innovation programme under grant agreement No 101000858 601 (TechOceanS), No 862626 (EuroSea), No 101081642 (OBAMA-NEXT). NEREA is 602 recognized as UN Ocean Decade Action No 64.2 under the OBON Program. We thank the WGIMT and WGZE of the International Council for the Exploration of the Sea (ICES) for facilitating this research. This study benefitted from collaboration with members of the Scientific Committee on Oceanic Research (SCOR) Working Group 157 (MetaZooGene). IDC acknowledge the support of NBFC (National Biodiversity Future Center) to Stazione Zoologica Anton Dohrn, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, “Dalla ricerca all’impresa”, Investimento 1.4, Project CN00000033. The authors are deeply grateful to the handling editor the manuscript, as well as the anonymous reviewers, for their invaluable comments and suggestions.
Author information
Authors and Affiliations
Contributions
IDC conceptualization and design; IDC, FL acquisition of data; IDC, FL GZ, RP analysis and interpretation of data; IDC, RP, FL, VR, YC drafting the manuscript. All authors significantly participated in critical revision of the manuscript draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Di Capua, I., Luise, F., Zampicinini, G. et al. Integrative approach to monitoring metazoan diversity and distribution in two Mediterranean coastal sites through morphology and organismal eDNA. Sci Rep 14, 19291 (2024). https://doi.org/10.1038/s41598-024-69520-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69520-2