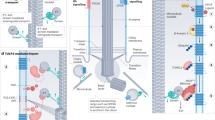
Abstract
Alterations to cilia are responsible for a wide range of severe disease; however, understanding of the transcriptional control of ciliogenesis remains incomplete. In this study we investigated whether altered cilia-mediated signaling contributes to the pleiotropic phenotypes caused by the Forkhead transcription factor FOXC1. Here, we show that patients with FOXC1-attributable Axenfeld–Rieger Syndrome (ARS) have a prevalence of ciliopathy-associated phenotypes comparable to syndromic ciliopathies. We demonstrate that altering the level of Foxc1 protein, via shRNA mediated inhibition, CRISPR/Cas9 mutagenesis and overexpression, modifies cilia length in vitro. These structural changes were associated with substantially perturbed cilia-dependent signaling [Hedgehog (Hh) and PDGFRα], and altered ciliary compartmentalization of the Hh pathway transcription factor, Gli2. Consistent with these data, in primary cultures of murine embryonic meninges, cilia length was significantly reduced in heterozygous and homozygous Foxc1 mutants compared to controls. Meningeal expression of the core Hh signaling components Gli1, Gli3 and Sufu was dysregulated, with comparable dysregulation of Pdgfrα signaling evident from significantly altered Pdgfrα and phosphorylated Pdgfrα expression. On the basis of these clinical and experimental findings, we propose a model that altered cilia-mediated signaling contributes to some FOXC1-induced phenotypes.
Similar content being viewed by others
Introduction
The primary cilium is a sensory organelle, present on most cells, that has essential roles in development and homeostasis. Mutation in genes encoding ciliary proteins, result in an extensive spectrum of phenotypes, in which severe congenital anomalies are over-represented. Consequently, such ciliopathies have been intensively investigated, both uncovering novel pathogenic mechanisms, and providing broader insight into the heritability of disease. For instance, ciliopathy inheritance patterns that were discordant with classical Mendelian models revealed the importance of tri-allelic inheritance1 and mutational load2, that in turn represent powerful paradigms for the causality of complex disease.
The importance of cilia stems from their role as a molecular nexus for signal transduction, with primary cilia mediating multiple pathways, including Hedgehog (Hh), and to a variable degree PDGFRα, TGF-β, Hippo and Wnt signaling3,4,5,6,7,8,9. Since cilia are indispensable to vertebrate Hh signaling, phenotypes indicative of Hh impairment represent clues that a disorder may be caused by ciliary dysfunction. Examples of such Hh-attributable developmental anomalies include alterations to digit number (syndactyly and polydactyly), the facial skeleton and the midline cerebellum10,11. Hydrocephalus, congenital heart disease and renal cysts exemplify additional cilia-associated phenotypes, that reflect roles in mechanosensation, chemosensation, and extracellular fluid movement12,13. However, clinical recognition of a ciliopathy can be obscured, by variable phenotypic severity and involvement of individual tissues. An additional complexity is provided by alterations to cilia positioning, that reflect perturbation of cellular orientation across tissue planes, or Planar Cell Polarity14.
Consistent with the premise that the mechanistic basis of a proportion of ciliopathies may be unrecognized, the majority identified to date are caused by mutations of major effect that induce severe and pediatric-onset disease. Milder alterations to cilia-mediated signaling would be expected to contribute to late-onset phenotypes; however, these remain largely unidentified. A second comparatively undefined area is the regulation of ciliogenesis. Only small numbers of transcription factors have been identified in vertebrates: including the Rfx gene family (Rfx1-4)15,16,17, and two Forkhead Box (FOX) family members (Foxj1 and Foxn4)18,19,20,21,22 that directly regulate development of motile cilia. FOX genes have fundamental physiological functions, that extend from angiogenesis and organ development, to cell cycle control23. Consequently, FOX gene mutations induce a diverse disease spectrum, that includes: autism, malignancy, immune deficiency, diabetes, stroke, and speech and language impairment24,25,26,27,28,29,30,31,32,33.
Our study concerns one intensively-studied FOX gene, FOXC1, which was originally identified as a regulator of organ development34. Subsequent studies revealed essential roles in arterial specification35, angio- and somitogenesis36,37,38, stem cell quiescence39,40 and hematopoietic progenitor formation41. Heterozygous FOXC1 mutation and copy number variation (segmental deletion and duplication) cause up to 50% of cases of Axenfeld–Rieger syndrome (ARS), a pediatric glaucoma-associated disorder whose variable systemic phenotypes include mid-facial hypoplasia, dental anomalies, congenital heart disease and auditory impairment42,43,44,45,46. In part guided by findings from the murine Foxc1ch/ch congenital hydrocephalus mutant34,47, and zebrafish morphants48, the disease spectrum has extended to include: cerebellar malformations, hydrocephalus, corneal angiogenesis, cerebrovascular disease, and multiple malignancies32,36,49,50,51,52. Involvement in stroke, primary open angle glaucoma, and the most severe subtype of breast cancer51,53, demonstrates FOXC1’s additional contribution to common late-onset diseases.
Intrigued that mutation, increased and decreased dosage of a single transcription factor could cause such pleiotropy, and recognizing that some phenotypes were indicative of a ciliopathy, we assessed the hypothesis that FOXC1 influenced ciliary function. Evaluation of cilia length and cilia-mediated pathways in vitro, together with studies in murine mutants, yielded evidence that support a model in which Foxc1 modulates aspects of ciliary function.
Results
Patient phenotypes induced by FOXC1 mutation or copy number variation
We first asked if there was clinical evidence that alterations to FOXC1 impaired cilia function. Patients with FOXC1-attributable Axenfeld–Rieger syndrome [mutation (n = 19), copy number variation (n = 22)]32,49,54,55 were evaluated for traits characteristic of altered cilia-mediated signaling. We established that the prevalence of ciliopathy-associated phenotypes was elevated, as illustrated by rates of midfacial hypoplasia (27%), congenital heart disease (27%), cerebellar hypoplasia (15%), hydrocephalus (8%) and others (Supplemental Table 1; Fig. 1). Several occurred at or above rates reported in ciliopathies: ventriculomegaly and hydrocephalus 19% compared to 23% in Joubert Syndrome, or congenital heart disease 27% compared to 5–18% in Nephronophthisis, Bardet–Biedl and McKusick–Kaufman Syndromes. Other phenotypes were less prevalent, as illustrated by a 15% prevalence of cerebellar hypoplasia, compared to ~ 100% in Joubert Syndrome56,57,58,59,60. Such findings may, in part, reflect the extreme heterogeneity of ciliopathies, as the prevalence of cerebellar hypoplasia (14%) in Bardet–Biedl Syndrome demonstrates61. A second factor may be that most syndromic ciliopathies are autosomal recessively inherited, with some cases of tri-allelic inheritance1, while ARS requires a single mutant allele. Amongst the observed phenotypes, polydactyly (Fig. 1A) is pathognomonic of altered Hh signaling, which specifies digit number in the developing limb bud62,63 where Foxc1 is expressed34. Equally, cerebellar anomalies reflect perturbation of the Hh signaling that is essential for cerebellar progenitor cell proliferation (midline cerebellar hypoplasia; Fig. 1B). Collectively, the clinical data demonstrate that FOXC1 mutation or dosage alteration induces variable multi-organ phenotypes, which coincide with the spectrum seen in cilia dysfunction.
Pleiotropic and variable phenotypes are consistent with ciliary dysfunction. (A) Skeletal phenotypes present in the cohort of patients with FOXC1 mutation or copy number variation include scoliosis and pre-axial polydactyly (arrowhead); the duplicated second phalanx of the left thumb was previously surgically removed. The mutation present in this individual (p.D117Tfs64) is predicted to result in loss of two thirds of the FOXC1 protein. (B) CNS phenotypes include cerebellar vermis hypoplasia, posterior fossa enlargement, and lateral ventricular dilation: example from an individual with a missense mutation (p.S82T). Images showing normal cerebellar and ventricular morphology, are outlined in green. (C) Ocular anomalies include irregular iris sphincter width (red arrow), posterior embryotoxon extending through 3 clock hours (green arrow), and asymmetric irides. Normal iris anatomy for comparison (green box).
Foxc1 expression influences length of primary cilia in fibroblast, renal and chondrogenic cell lines
We next employed three mammalian cell lines, to determine whether manipulation of the level of Foxc1 impacted cilia structure. Since ARS is frequently caused by segmental duplication and deletion that increase and decrease the copy number of FOXC1, the effects of Foxc1 overexpression, and inhibition, were evaluated in vitro. In murine fibroblasts (NIH3T3), four independent Foxc1-targeting shRNAs reduced protein levels of Foxc1 by 40–61% and induced mild cilia shortening relative to cells expressing either a non-targeting shRNA or control plasmid (Fig. 2A,J). Comparable cilia lengthening was observed with stable overexpression of Foxc1 (Fig. 2A). To better evaluate the magnitude of these effects, automated image analysis methodology was developed, and validated by comparing manual and automated cilia length measurements in cells treated with a Foxc1-targeting shRNA or vector control (pLKO.1). The mean cilia lengths quantified by the two approaches, were concordant (automated: Foxc1 shRNA 0.85, control 1.0, P = 4.5 × 10−4; manual: Foxc1 shRNA 0.87, P = 1.6 × 10−3; Supplemental Fig. 1). Measurement of a larger number of cilia revealed that Foxc1 inhibition is accompanied by an altered cilia length distribution in the cell population (PKW = 7 × 10−3; n = 7431 cilia; Fig. 2B), with an increased proportion of short cilia (shRNA 34–44%; controls 24–27%; P = 1.7 × 10−4; Fig. 2C; Supplemental Fig. 1C for methodology), and overall, a 13% shortening relative to controls (6–19% for individual shRNAs; Fig. 2C, Supplemental Fig. 5). Increased Foxc1 expression induced a 9% lengthening, primarily due to an increased subpopulation of cells with longer cilia (Foxc1 overexpression 58%; controls 50%; P = 0.02; n = 9590 cilia; Fig. 2C). These data illustrate that automated analysis in large numbers of cells readily resolves mild alterations in cilia length. Quantification of cilia length in a second murine cell line that expresses Foxc1 at a higher level [inner medullary collecting duct cells (IMCD3)], revealed analogous alterations (Foxc1 shRNA inhibition: 30–31% shortening, P = 7.6 × 10−7; overexpression: 10% lengthening, P = 6.5 × 10−5, Fig. 2D–F). These data are corroborated by use of two constructs that lack the transcription factor’s C-terminal activation domain, while the frameshift mutation truncates Foxc1’s DNA binding domain [p.E231* and p.N129Tfs*52]. Expression of either transcriptionally inactive construct, failed to induce changes to cilia length (Supplemental Fig. 2). In a third, and chondrogenic cell line (ATDC5), CRISPR/Cas9 mutagenesis of Foxc1 induced an 11% reduction in cilia length compared to CRISPR-treated control cells (P = 3 × 10−5; Fig. 2G–I). This demonstrates that a targeted loss of function Foxc1 mutation, recapitulates the effect of shRNA inhibition observed in NIH3T3 and IMCD3 cells, with ~ 2 fold changes in Foxc1 protein levels (Fig. 2J).
Altered levels of Foxc1 are associated with changes to cilia length in three cell types. (A) Representative images of Arl13b/γ-tubulin ciliary staining in NIH3T3 cells expressing a Foxc1-targeting shRNA, or with Foxc1 overexpression (OE) (non-targeting shRNA control: non-trg.; vector controls: pLKO.1, pLXSH). These reveal shortening of the cilium (red) with shRNA inhibition, and the converse with OE. (B) Quantification of cilia length as fold change relative to vector controls from independent experiments (shRNA: n = 6; OE: n = 7). (C) Prevalence of short (blue bars; length ≤ lower quartile of controls) and long cilia (green bars; length ≥ median of controls); see Supplemental Fig. 1 for details. (D–F) In IMCD3 cells, Foxc1 knock-down and OE respectively reduce and increase cilia length; graphs depict fold change in cilia length and prevalence of short and long cilia (shRNA, OE: n = 1). (G–I) In ATDC5 cells, the decreased cilia length induced by CRISPR mutagenesis of Foxc1 is attributable to an increased proportion of cells with short cilia (n = 4). (J) In NIH3T3 cells, Foxc1 protein levels (relative to WT) are reduced ~ 2 fold (40–61%) by Foxc1-targeting shRNAs (n = 1). [Statistical analyses: boxplots—Dunn’s (post hoc Kruskal–Wallis) test; barplots—nested ANOVA (C shRNA), one-way ANOVA (C Foxc1 OE, I)].
Altered Foxc1 dosage impacts cilia-mediated Hh signaling
To test whether the alterations in cilia length were associated with perturbed cilia-dependent signaling, we first assayed the activity of the Hedgehog pathway, that is mediated by the primary cilium in most vertebrate tissues64,65,66. The Gli proteins (Gli1–Gli3) represent key effectors of the pathway, and the level of Gli1 was used as the initial readout. NIH3T3 cells with shRNA-mediated knockdown of Foxc1 accumulate lower levels of Gli1 on stimulation of Hh signaling with Smoothened agonist (SAG; Fig. 3A–C). Equally, cells overexpressing Foxc1 exhibit increased levels of Gli1 without the application of SAG (Fig. 3D). To validate these findings, we next assayed the effect of manipulating Foxc1 levels using a second cell line: immortalized embryonic primary chondrocytes that express high levels of endogenous Foxc1 (Supplemental Fig. 3). In these mesenchymal-derived cells, shRNA-mediated knockdown of Foxc1 significantly decreased Gli1 expression (Fig. 3E). Evidence that these alterations are not confined to Gli1, was provided by the significantly increased expression of Gli2 (the main activator of mammalian Hh signaling) and Ptch1 observed in Foxc1-overexpressing fibroblasts (Fig. 3F). Furthermore, overexpression of Foxc1 results in more rapid and increased Gli1 protein expression in serum-starved cells in response to SAG stimulation (Fig. 3G). Taken together, these data demonstrate that alterations to the level of Foxc1 perturb the in vitro expression of major Hh pathway components.
Altered levels of Foxc1 induce aberrant ciliary Hedgehog signaling. (A–D) In NIH3T3 cells, quantitative Western Immunoblots demonstrate that shRNA inhibition of Foxc1 decreases the basal level of Gli1 expression, while Foxc1 overexpression has a converse effect. (B,C) Reduced Foxc1 expression leads to twofold decrease in Gli1 protein levels [measured in serum-starved cells stimulated with 400 nM Smoothened agonist (SAG), for 20 h; n = 1 for each of the four shRNAs]. (D) Increased Foxc1 expression results in ~ 2 fold increase in the basal level of Gli1 protein (n = 4). (E) In immortalised E16.5 chondrocytes, that express high levels of Foxc1, Foxc1 shRNA inhibition decreased basal level of Gli1 mRNA (n = 3). (F) Cells overexpressing Foxc1 exhibit increased basal levels of Gli1, Gli2 and Ptch1 mRNA (n = 3). (G) Immunoblots demonstrate that overexpressing Foxc1 induces faster accumulation of Gli1 protein in serum-starved NIH3T3 cells. Quantification reveals significantly increased Gli1 protein levels for Foxc1 OE at 3 and 6 h (n = 3). [Statistical analyses: Tukey HSD test post hoc one-way (A–F) or two-way ANOVA (G)].
Foxc1 induces accumulation of Gli2 at the ciliary tip in vitro
A notable feature of vertebrate Hh signaling is that components change subcellular localization in response to ligand activation. This is illustrated by the accumulation of Gli2 at the axonemal tip67, which is essential to signal transduction. Accordingly, we established a clonal NIH3T3 cell line stably expressing Gli2-mGFP, and first demonstrated that it recapitulated the SAG-dependent accumulation of Gli1 protein observed in NIH3T3 cells (Fig. 4A,B). The elevated basal level of Gli1 protein observed in these Gli2-mGFP cells, supports a cooperative effect of Foxc1 and Gli2 proteins on Hh pathway output (Fig. 4A–C). Overexpression of Foxc1, increased Gli2-mGFP signal intensity at the ciliary tip by 92% (PKW = 8.2 × 10−65; n = 4231 measurements). Stimulation with SAG further increased signal intensity ~ 2 fold (PKW = 6.2 × 10−47; Fig. 4B,C) and such enhanced axonemal tip accumulation of Gli2 is consistent with Foxc1 impacting a core cilia-mediated signaling pathway.
Foxc1 alters the dynamics of Hh signaling and enhances Gli2 accumulation at the ciliary tip. (A) Immunoblots demonstrate that overexpressing Foxc1 induces faster accumulation of Gli1. Quantification in serum-starved Gli2-mGFP-expressing NIH3T3 cells reveals substantially higher Gli1 protein levels with Foxc1 OE: 14-, 20- and 26-fold increases vs non-treated wild-type control at 0, 3 and 6 h (n = 1). Note the progressive accumulation of Gli1 protein in response to SAG treatment. (B) Images illustrate the increased accumulation of Gli2-mGFP at the axonemal tips of Foxc1 OE cells, including after stimulation with SAG. Orientation of primary cilia, from basal body (γ-tubulin staining) to axonemal tip (Gli2-mGFP), is depicted by the red arrow. (C) Axonemal tip Gli2-mGFP signal intensity, expressed as fold change relative to pLXSH control, is significantly increased by Foxc1 OE. The prevalence of high and low Gli2-mGFP signal intensity in 7 independent experiments is also significantly altered compared to control [high Gli2-mGFP signal intensity ≥ upper quartile, green bar; low ≤ lower quartile of control, blue]. [Statistical analysis: boxplots—Dunn’s (post hoc Kruskal–Wallis) test; barplots—Tukey HSD post hoc one-way ANOVA].
Altered levels of Foxc1 impact platelet-derived growth factor signaling
Since several receptor tyrosine kinase signaling pathways are partially mediated by cilia, we next assayed the PDGFRα pathway to determine whether Foxc1’s effects on cilia-dependent signaling extend beyond Hedgehog signal transduction. Initiated at primary cilia under conditions of serum starvation, ligand stimulation by platelet-derived growth factors (PDGF-A to C) induces phosphorylation of specific PDGFRα tyrosine residues, such as pY754, that provide a readout of receptor activity. We observed that serum-starved NIH3T3 cells accumulate lower levels of PDGFRα protein with either knockdown or overexpression of Foxc1 (individual shRNA range 0.44–0.76, OE 0.63, control 1.0; P < 0.004; Fig. 5). When stimulated with cognate PDGF-AA ligand, increased and decreased levels of Foxc1 lead to reduced kinase activity of the PDGFRα receptor. Foxc1 shRNA knockdown induced a two-fold reduction in levels of pY754 autophosphorylated PDGFRα (P = 4.5 × 10−5), while Foxc1 overexpression resulted in a 3 fold reduction (P = 3 × 10−7; Fig. 5D–F). These data demonstrate that Foxc1 moderately affects protein levels of total PDGFRα while strongly impacting its ligand-dependent phosphorylation. In the context of the Hh data (Figs. 3, 4), these results demonstrate that in vitro, altered levels of Foxc1 affect two cilia-mediated signaling pathways.
Altered levels of Foxc1 impact PDGFRα signaling. (A–C) Foxc1 shRNA inhibition significantly reduced the level of total Pdgfrα, and auto-phosphorylated pY754-Pdgfrα in serum-starved NIH3T3 cells. This effect was observed both in the presence and absence of Pdgf-AA ligand stimulation (n = 1 for each of the four shRNAs). (D–F) Foxc1 overexpression induced comparable reductions in total Pdgfrα, and pY754-Pdgfrα levels, indicative of impaired cilia-mediated Pdgf signaling (n = 3). [Statistical analyses: Tukey HSD post hoc one-way (B,C) or two-way (E,F) ANOVA. Statistical analysis in panels (B) and (C) shows data pooled for all shRNAs].
Foxc1 mutation impacts meningeal ciliation
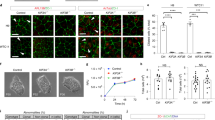
To establish if comparable changes occurred in vivo, embryos derived from two mutant strains with well-characterized Foxc1 null alleles34,68 were used: Foxc1Δ/Δ (Cre-mediated deletion of the Foxc1 open reading frame) and Foxc1−/− (Foxc1lacZ deletion). As expected, the generated homozygous mutant embryos displayed severe dilation of the lateral ventricles (data not shown). To assess whether loss of Foxc1 influences cilia length in vivo, primary cultures of meningeal cells were established and the length of primary cilia determined. The mean cilia length in meningeal cells from heterozygous and homozygous embryos was reduced (Foxc1Δ/+ 2.63 μm (8% reduction), Foxc1Δ/Δ 2.56 μm (10% reduction), WT 2.85 μm; n = 8972 cilia; P < 0.04, Fig. 6). The altered cilia length distribution, evident from a reduced proportion of longer cilia and increased proportion of shorter cilia (Fig. 6B), recapitulates the changes observed in vitro (NIH3T3, IMCD3 and ATDC5 cells; Fig. 2).
Reduced cilia length in meningeal cells cultured from Foxc1 mutant embryos. (A) Representative images of Arl13b/γ-tubulin staining in primary meningeal cell cultures from Foxc1ΔTg(EIIa-cre) and wild-type sibling E15.5 embryos. Note the shortening of the cilium (red), most apparent in the homozygous mutant (Foxc1ΔΔ) compared to controls. (B) Quantification of cilia length in primary cultures of meninges isolated from 4 litters comprising 6 Foxc1+/+, 16 Foxc1+/Δ and 5 Foxc1Δ/Δ embryos. (C) Mean cilia length is reduced by 8% and 10% in Foxc1+/Δ and Foxc1Δ/Δ relative to wild-type cultures. This is attributable to a decreased proportion of longer cilia, as demonstrated by the barplot on the right depicting the prevalence of short (blue bars; length ≤ lower quartile of controls) and long cilia (green bars; length ≥ median of controls). [Statistical analysis: one-way ANOVA].
Foxc1 mutation dysregulates meningeal expression of Hedgehog and PDGFRα signaling components
In the CNS, Foxc1 is primarily expressed in the meninges, a complex tissue that represents an essential source of signaling molecules for the developing brain, and skull49,69,70. The diverse signaling crosstalk extends from retinoic acid and BMP signaling to FGFs and others; from these we selected two ciliary-mediated pathways for analysis. Immunoblots of E14.5 forebrain meningeal lysates, pooled by genotype, demonstrated Gli1 protein expression was strongly depleted in Foxc1+/− heterozygotes, with near complete loss of expression in homozygous mutants (Fig. 7A). This reduction in a key output of Hh signaling, was accompanied by 3 fold elevation in the level of the repressor form of Gli3, and increased protein expression of Sufu (Fig. 7B,C). Since these data demonstrate dysregulation of Hedgehog signaling in embryonic meninges, we proceeded to quantify meningeal PDGFRα protein expression at the same timepoint. Immunofluorescent staining demonstrated ~ 1.9-fold reductions in both total PDGFRα, and the level of the active autophosphorylated form, in E14.5 Foxc1−/− embryos compared to wild type littermates (PDGFRα: Foxc1−/− 0.55, WT 1.0, P = 2.6 × 10−4; pY754-PDGFRα: Foxc1−/− 0.54, WT 1.0, P = 0.048; Fig. 8A–G). Immunoblot analysis of meningeal PDGFRα protein levels revealed similar reductions to those observed by immunofluorescent analysis (Fig. 8H). These in vivo data for Hh and PDGFRα signaling, accord with the in vitro results.
Dysregulated Hedgehog signaling in the meninges of Foxc1 mutant embryos. (A–C) Western immunoblot analysis demonstrates that meningeal expression of core Hh components is dysregulated by Foxc1 mutation. (A) Gli1 protein expression is decreased in Foxc1+/Δ heterozygotes, and almost entirely lost in Foxc1Δ/Δ meninges. (B) Foxc1 mutation is associated with increased expression of full length Gli3, and its increased processing into the C-terminally truncated repressor form (Gli3R). (C) Expression and phosphorylation of Sufu, a core Hh pathway inhibitor, is also increased with Foxc1 mutation. [Quantification above each gel: fold change in signal, normalized to actin, relative to wild-type. Samples pooled from n = 18 embryos (7 Foxc1+/+; 8 Foxc1+/Δ, 3 Foxc1Δ/Δ].
Decreased Pdgfrα expression in the meninges of Foxc1 mutant embryos. (A) At E14.5, Pdgfrα immunofluorescence is decreased in the forebrain meninges of Foxc1-null embryos. (B) This is particularly apparent in the meninges lining the anterior cranial fossa [insets from (A)]. (C,D) The mean Pdgfrα signal intensity in the forebrain meninges is decreased by 50% (P = 2.6 × 10−4). Consistently, RNA-sequencing demonstrates significantly reduced meningeal Pdgfra mRNA expression [27% mean FPKM decrease, q = 6.4 × 10−5], while in contrast Pdgfrb expression is unaltered. (E) E14.5 Foxc1−/− embryos exhibited comparable reductions in forebrain meningeal pY754 Pdgfrα expression. (F) Quantification confirms the significant decrease in the mean signal intensity of the active phosphorylated form of Pdgfrα, and comparable reductions in the level of the tight junction protein Zo1, relative to wild-type embryos. (H) Western immunoblots of meningeal tissue lysates reveal similar reductions in total Pdgfrα levels to those observed by immunofluorescence (replicates as in Fig. 7). [Panels 8C, F, G: 6 Foxc1+/+, 6 Foxc1−/− embryos; 8D: 5 Foxc1+/+, 4 Foxc1Δ/Δ embryos]. [Mgs meninges, LV lateral ventricle, ChP choroid plexus, FC mesenchymal condensation forming falx cerebri. Statistical analyses: one-way ANOVA; FPKM Fragments Per Kilobase of transcript, per Million mapped reads].
Integration with independent gene expression resources
RNA sequencing of meningeal tissue from E14.5 Foxc1−/− and wildtype embryos revealed the dysregulation of multiple cilia-associated pathways, including Hh, Wnt, TGF-β and PDGFR (Supplemental Fig. 4A–F). In addition to recapitulating findings of altered meningeal Hh and PDGFRα signaling (Figs. 7 and 8), RNA sequencing demonstrated that the effect on PDGF signaling was specific to Pdgfra, since Pdgfrb mRNA expression was unaffected (Supplemental Fig. 4E; Fig. 8D). We then compared transcriptional changes in our dataset to published RNA expression data derived from Pdgfc−/−/PdgfraGFP/+ meninges: double mutants with profoundly altered PDGFRα signaling, whose abnormal meningeal and cerebellar development phenocopies Foxc1−/− embryos. Analysis revealed strong correlation of changes in expression of the 1473 significantly dysregulated genes in the meninges of Pdgfc−/−/PdgfraGFP/+ mice, and those also expressed in Foxc1Δ/Δ embryonic meninges (ρ = 0.77, 95% CI 0.75–0.79, P = 2.2 × 10−16, Fig. 9A,B). Concordant up- and down-regulated expression, is particularly evident at the top and bottom of the gene set (Fig. 9C); 90% of the genes expressed in the meninges of both mutants are significantly dysregulated in both datasets (1193 of 1324 genes). These findings indicate that multiple genes dysregulated by Foxc1 mutation in embryonic meninges comprise PDGFRα signaling targets.
Similar transcriptional changes in the meninges of Foxc1 and Pdgf pathway mutant mice. (A) Experimental design for analysis of differentially expressed genes in the meninges of Foxc1Δ/Δ embryos (4 Foxc1Δ/Δ, 5 Foxc1+/+) and mice with severely perturbed Pdgfrα signaling (Pdgfc−/−; PdgfraGFP/+). (B) The majority of genes dysregulated in the Pdgf mutant are similarly up- or down-regulated in Foxc1 mutant embryos; a correlation that extends to 1193 of 1324 genes; correlation coefficient ρ = 0.77, P = 2.2 × 10−16. (C) Restricting analysis to the 50 most upregulated, and 50 most downregulated genes, makes the similar differential expression evident: more than 80% of these most dysregulated genes in Pdgfc−/−; PdgfraGFP/+ mice are also significantly dysregulated in the meninges of Foxc1Δ/Δ embryos.
Discussion
This study presents evidence that mutation of Foxc1 impacts cilia function. The identification in FOXC1-attributable Axenfeld–Rieger cases of an increased prevalence of ciliary-associated phenotypes, led to the premise that ciliary dysfunction may contribute to this heterogeneous, multi-organ phenotype. We found evidence supporting this hypothesis in vitro, where shRNA inhibition and overexpression of Foxc1 induce reciprocal effects on cilia length. Comparable changes occurred in Foxc1+/Δ and Foxc1Δ/Δ embryonic meninges, with the greatest reductions observed in homozygous mutants. The cilia length changes were associated with dysregulated Hh and Pdgfrα signaling, and analyses of embryonic meninges—a source of extrinsic signals that regulate cortical neurogenesis and that strongly expresses Foxc1—directly recapitulated these in vitro results. Since Foxc1 mutation is known to disrupt meningeal retinoic acid synthesis71,72, the presented data demonstrate dysregulation of additional pathways, with alterations that extend beyond transcriptional outputs (Gli1 and Gli3) to include a core inhibitor of Hh pathway activity (Sufu)73. Together with the perturbed partitioning of Gli2 protein observed within the cilium, these results reveal altered Foxc1 expression perturbs different facets of Hh signaling. The substantial alterations to cilia-mediated signaling with comparatively mild changes in cilia length, have parallels to the visibly normal cilia but Hh-related patterning defects, or loss of PDGFRα signaling, observed with some Bardet–Biedl Syndrome mutations74,75,76,77.
As one of the first receptor tyrosine kinases determined to localize to primary cilia, the concordant in vitro and ex vivo PDGFRα signaling data are equally important. They demonstrate Foxc1’s influence on a second cilia-mediated signaling pathway. Ligand binding to PDGFRα receptors in the ciliary membrane induces receptor dimerization, autophosphorylation, and activation of Ras, P13K-AKT and MAPK pathway components. The significantly altered PDGFRα expression and activity (pY754 phosphorylation), with unchanged Pdgfrb expression, demonstrates effect specificity in the PDGF signaling network. PDGFRα signaling has well characterized roles that encompass formation of neural crest derived tissues, and multiple aspects of CNS development78,79,80. As seen with Foxc1, targeted disruption of Pdgfra induces embryonic lethality; and Pdgfrα mutants exhibit craniofacial defects, cardiac outflow tract malformations and somite patterning defects that are characteristic of Foxc1 mutation79,81. To determine whether a common mechanism accounted for these similarities, genes differentially expressed in the meninges of Foxc1 and Pdgf pathway mutant mice were compared. The prominent correlation between the datasets (ρ = 0.77, P = 10−16) with 1193 of 1324 dysregulated genes common to both mutants, are consistent with Foxc1 controlling part of a network of PDGF-responsive genes via effects on the PDGFRα receptor.
The data presented also provide a clinical explanation for the pleiotropic phenotypes associated with FOXC1 mutation. Pleiotropy is an established feature of ciliopathies, and can result in a spectrum that extends from involvement of individual tissues to diverse combinations of organs82. Consistently, the Foxc1 murine mutant congenital hydrocephalus was originally reported as one of the earliest examples of a pleiotropic mutation variably altering a large number of organ systems34,47. The ARS phenotype most suggestive of a ciliary component was cerebellar hypoplasia49, which reflects the reliance of cerebellar granule cell precursor proliferation on correct levels of Hh signaling70,83. Multiple other anomalies (Supplemental Table 1) are prevalent in the ciliopathy spectrum, consistent with the premise that ciliary dysfunction may contribute to such ARS phenotypes. Potentially, there may be some relevance to late onset disorders because Hh signaling’s maintenance of vascular integrity84,85, and pericyte recruitment84,85,86,87, offer a plausible explanation for FOXC1's involvement in cerebral small vessel disease and stroke32.
A limitation of this study is that our understanding of the mechanisms by which Foxc1 influences cilia function remain incomplete. The primary effects observed were to cilia-dependent Hh and Pdgfrα signaling, with relatively mild changes to cilia length. Indeed, the RNA sequencing experiment assaying developmental signaling in E14.5 meninges, did not resolve transcriptional changes involving cilia formation or maintenance. So the results observed may reflect direct effects from loss of Foxc1 on these two signaling pathways. In this context, it is intriguing that data from studies independently deleting intraflagellar transport proteins from the murine neural crest also associated primary cilia with ARS. The first demonstrated that ablating cilia by conditional Ift88 deletion induced ocular mal-development characteristic of ARS88: findings reiterated by a second that targeted Ift4689. Such phenocopying of FOXC1’s phenotypes by deleting different IFT proteins supports a link between cilia and the pathogenesis of ARS, and will encourage future studies to define the precise mechanisms.
In conclusion, we have shown that alterations to the level of Foxc1 affect cilia length in vitro and ex vivo, and impact the function of two cilia-associated signaling pathways. The findings increase the number of Forkhead genes and clades implicated in some facet of cilia biology, in turn supporting sub-functionalization across a larger proportion of this intriguing transcription factor family. Overall, we propose the model that altered cilia-mediated signaling contributes to FOXC1’s pleiotropic phenotypes.
Methods
Plasmids, antibodies and other reagents
Retroviral plasmids for stable expression of non-tagged mouse Foxc1 ORFs was created by InFusion HD® subcloning into pLXSH vector and QuickChange II site-directed mutagenesis. mGFP-Gli2-pCEFL vector for expression of mouse Gli2 N-terminally tagged with monomeric GFP (mGFP) was a gift from Dr. Philip Beachy (Stanford University School of Medicine, Stanford, CA). Lentiviral shRNA plasmids from TRC1.5 mouse MISSION® shRNA libraries were obtained from the RNAi Screening Core (Li Ka Shing Institute of Virology, University of Alberta). Detailed information on all ORF and shRNA vectors, complete list of the antibodies, primers, as well as other reagents used in the study are provided in Supplemental methods (Tables S2–S6).
Cell lines
NIH3T3 mouse fibroblasts and derivative cell lines were grown at in DMEM supplemented with 10% FBS; mIMCD3 inner medullary collecting duct cells—in DMEM:F12 supplemented with 10% FBS; ATDC5 chondrogenic cells and derivative lines—in DMEM:F12 supplemented with 5% FBS and 2 mM GluaMAX-I. Immortalised mouse chondrocytes were grown in DMEM with 10% FBS. All cell lines were kept at 37 °C in 5% CO2, in growth media containing 100 U/ml penicillin and 100 μg/ml streptomycin. Clonal NIH3T3 cell line stably expressing Gli2-mGFP was generated by transfection of parental cells with subsequent selection in 800 μg/ml Genetecin. NIH3T3, Gli2-mGFP NIH3T3 and mIMCD3 pools of cells stably expressing Foxc1 were made by transduction with Foxc1 ORF retroviral particles followed by selection in 100–500 μg/ml Hygromycin. Selection of NIH3T3 and mIMCD3 pools of cells transduced with lentiviral shRNA particles was performed in 2.5 µg/ml Puromycin. Clonal ATDC5 cell line with mutation to Foxc1 (Foxc1c.345_355del/c.353_356del) was generated using Alt-R Crispr-Cas9 system (IDT) as previously described90. Additional details provided in Supplemental methods.
Measurement of cilia length in vitro
Measurement of cilia length in NIH3T3 cells was performed after 20 h starvation in DMEM without FBS. In brief, after fixation in Dent’s solution, cilia and basal bodies of cells were immunofluorescently labelled using anti-Arl13b and anti-γ-tubulin antibodies respectively (see Supplemental methods for details). Images were collected using Zeiss LSM 700 laser scanning confocal microscope and subjected to quantification of cilia length with automatic Cell Profiler91 (the Broad Institute, Cambridge, MA) software-based image analysis pipeline. Measurements of cilia length in ATDC5 (automatic, Cell Profiler) and mIMCD3 (manual, using Fiji software92) were performed under conditions of growth in complete media with 10% FBS. For each cell line, these absolute measurements (μm), were then plotted graphically, relative to the mean cilia length of the appropriate control for each condition (pLKO.1 for shRNAs; pLXSH for overexpression). For analyses of cilia length, short cilia were defined as having a length ≤ lower quartile of controls, and long cilia as ≥ median of controls.
qPCR analyses
Total RNA was isolated with RNeasy Plus Mini Kit (Qiagen), quantified and used for cDNA synthesis with Primescript RT Master Mix (Clontech). qPCR reactions were run with SYBR® Premix Ex Taq (Tli RNAse H Plus) master mix (Clontech) on LightCycler® 96 Instrument and analysed using LightCycler® 96 Application (Roche Life Science). Primer sets used are provided in Supplemental methods (Table S6).
Quantitative western blotting
Embryos were harvested at E14.5 and dissected in ice-cold TBS. Isolated tissue was snap-frozen in a dry ice bath and stored and stored at − 85 °C until further processing. Cells and isolated tissue were lysed in 1.5% SDS lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1.5% SDS) supplemented with protease/phosphatase inhibitor cocktail (1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin or Halt Protease Inhibitor Cocktail (Thermo); 0.5 mM Na3VO4, 5 mM NaF, 10 mM β-glycerophosphate) and passed through QIAshredder columns (Qiagen). Obtained protein samples were normalised using BCA Protein Assay Kit (Thermo), resolved by SDS-PAGE (NuPage 4–12% Bis–Tris gels, Invitrogen), transferred to Immobilon-FL PVDF membranes (EMD Millipore) and blocked with Odyssey® TBS Blocking Buffer (Li-Cor). Next membranes were incubated with relevant primary antibodies, followed by IRDye-conjugated secondary antibodies. Resulting membranes were scanned with Odyssey® Imaging System (Li-Cor). Protein levels were quantified using Odyssey® Application Software (Li-Cor), normalised to actin, and expressed as fold change of a relevant control. List of primary and secondary antibodies used can be found in Supplemental methods (Table S4).
Measurement of ciliary Gli2 accumulation
Accumulation of Gli2 at the cilia tips was measured in NIH3T3 Gli2-mGFP cells following 20 h starvation in DMEM medium without FBS, and subsequent stimulation with Smoothened agonist (SAG). After fixation in Dent’s solution, cells were immunofluorescently labelled using anti-Arl13b and anti-γ-tubulin antibodies respectively (see Supplemental methods for details). Images were collected using Zeiss LSM 700 laser scanning confocal microscope and quantified with automatic Cell Profiler (99) software-based image analysis pipeline.
Experimental animals
The Foxc1−/− embryos carrying the null Foxc1lacZ mutation were generated and genotyped as previously described34,93. The embryos carrying Foxc1Δ/Δ null mutation were generated by deletion of the entire Foxc1 open reading frame by crossing Foxc1 flox/flox mice to EIIa-cre mice as described previously68. Embryonic age was determined by defining noon on the day of vaginal plug as E0.5. Genotyping of embryos was performed by PCR as described before34 and additionally confirmed by qPCR end-point genotyping. Primers used for qPCR genotyping are provided in Supplemental methods (Table S6).
Primary culture of murine meningeal cells
Embryos were harvested at E15.5 in ice-cold HBSS with calcium and magnesium. Upon removal of skin and cranium, forebrain leptomeningeal tissues were peeled off. Tissues were incubated in digestion solution consisting of DMEM media supplemented with 2% BSA and 5 mg/ml collagenase II (Worthington Biochemical, NJ) with occasional vortexing, for 3 h at 37 °C until dispersed. Released cells were gently centrifuged, washed once in complete DMEM media supplemented with antibiotic-antimitotic solution (100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B; Thermo) and 10% FBS. Cells were resuspended, passed through μm cell strainer to remove aggregates, and grown in complete DMEM. After 5 days in culture, samples were fixed in Dent’s solution and immunofluorescently labelled with anti-Arl13b and anti-γ-tubulin antibodies (see Supplemental methods for details). Automatic quantification of cilia length from confocal microscopy images was performed with Cell Profiler91.
Immunostaining of mouse tissues
Embryos were harvested at E14.5 in cold PBS and fixed in 4% paraformaldehyde (PFA) for 4 h at 4 °C. Following fixation embryos were equilibrated in 20% sucrose in PBS, embedded in Clear Frozen Section Compound (VWR, Richmond, IL) and frozen in a dry ice bath. Coronal cryosections (12–16 μM) thick were made using Leica CM1900, CM1950 cryostats and stored at − 85 °C. PDGFRα signalling in the meninges was assayed using anti-PDGFRα and anti-pY754 PDGFRα antibodies. Tight junctions in the meninges were labelled with anti-Zo1-AF555 antibody conjugate. Images were collected using Zeiss LSM 700 laser scanning confocal microscope or Zeiss Axioscan Z1 slide scanner. Measurements of PDGFRα and pY754 PDGFRα signal in the meninges were done with Fiji92.
RNA sequencing and gene expression datasets
Foxc1Δ/Δ and wild-type embryos were harvested at E14.5 in cold PBS and dissected in RNALater solution (Thermo). RNA was isolated from meningeal tissue by RNeasy Mini Kit (Qiagen) and sequenced at the BRC Sequencing Core (University of British Columbia, Vancouver, BC; see Supplemental methods for details). These data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE227703 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE227703). The dataset of transcriptional changes in the meninges of Pdgfc−/−; PdgfraGFP/+ mice was obtained from NCBI’s Gene Expression Omnibus (GEO accession: GSE67644)94.
Statistics
Statistical analyses were performed using RStudio version 0.99 software (RStudio Inc) running R language version 3.4.2 (The R Project for Statistical Computing). Analyses of statistical significance (P < 0.05) were performed with one-way ANOVA, two-way ANOVA, Tukey HSD test post hoc one-way and two-way ANOVA, nested ANOVA or Dunn’s (post hoc Kruskal–Wallis) test, as indicated for specific experiments. Bar plots show mean values ± SEM. Box-whisker plots show quartiles, median (black lines) and mean (black dots) values. Significance codes ***P < 0.001, **P < 0.01, *P < 0.05.
Patient cohort and phenotyping
A collaborative effort encompassing clinicians and researchers at multiple institutions, identified 41 individuals with FOXC1 mutation or copy number variation. For one member of this cohort, medical records and MRI data were provided remotely; the other participants were phenotyped at the University of Alberta. 18 of these 40 primarily local patients had been included in a previous study investigating cerebrovascular disease32.
Study approval
Ethical approval was provided by the University of Alberta Health Research Ethics Board, with written informed consent received from all participants prior to their inclusion in the study. All experiments were performed in accordance with the relevant guidelines and regulations.
Animal experiments were approved by the IACUC of the University of Alberta. All experiments were performed in accordance with relevant guidelines and regulations. This study is reported in accordance with ARRIVE guidelines.
Data availability
All data generated and/or analysed in this study are available from the corresponding author upon reasonable request. The RNA-sequencing data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE227703 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE227703).
References
Katsanis, N. et al. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science 293(5538), 2256–2259 (2001).
Zaghloul, N. A. et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet–Biedl syndrome. Proc. Natl. Acad. Sci. USA 107(23), 10602–10607 (2010).
Huangfu, D. et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426(6962), 83–87 (2003).
Schneider, L. et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 15(20), 1861–1866 (2005).
Simons, M. et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat. Genet. 37(5), 537–543 (2005).
Habbig, S. et al. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 193(4), 633–642 (2011).
Clement, C. A. et al. TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 3(6), 1806–1814 (2013).
Christensen, S. T. et al. Primary cilia and coordination of receptor tyrosine kinase (RTK) and transforming growth factor beta (TGF-beta) signaling. Cold Spring Harb. Perspect. Biol. 9(6), a028167 (2017).
Schwarz, H. et al. Biallelic ANKS6 mutations cause late-onset ciliopathy with chronic kidney disease through YAP dysregulation. Hum. Mol. Genet. 31(9), 1357–1369 (2022).
Wechsler-Reya, R. J. & Scott, M. P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22(1), 103–114 (1999).
Chiang, C. et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383(6599), 407–413 (1996).
Waters, A. M. & Beales, P. L. Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 26(7), 1039–1056 (2011).
Tobin, J. L. & Beales, P. L. The nonmotile ciliopathies. Genet. Med. 11(6), 386–402 (2009).
Butler, M. T. & Wallingford, J. B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell. Biol. 18(6), 375–388 (2017).
Swoboda, P., Adler, H. T. & Thomas, J. H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5(3), 411–421 (2000).
Haycraft, C. J. et al. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 128(9), 1493–1505 (2001).
Choksi, S. P. et al. Switching on cilia: Transcriptional networks regulating ciliogenesis. Development 141(7), 1427–1441 (2014).
Chen, J. et al. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Investig. 102(6), 1077–1082 (1998).
Yu, X. et al. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40(12), 1445–1453 (2008).
Stubbs, J. L. et al. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40(12), 1454–1460 (2008).
Campbell, E. P., Quigley, I. K. & Kintner, C. Foxn4 promotes gene expression required for the formation of multiple motile cilia. Development 143(24), 4654–4664 (2016).
Newton, F. G. et al. Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization. Dev. Cell 22(6), 1221–1233 (2012).
Golson, M. L. & Kaestner, K. H. Fox transcription factors: From development to disease. Development 143(24), 4558–4570 (2016).
Mariani, J. et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162(2), 375–390 (2015).
Pantelouris, E. M. Absence of thymus in a mouse mutant. Nature 217(5126), 370–371 (1968).
Schuddekopf, K., Schorpp, M. & Boehm, T. The whn transcription factor encoded by the nude locus contains an evolutionarily conserved and functionally indispensable activation domain. Proc. Natl. Acad. Sci. USA 93(18), 9661–9664 (1996).
Bennett, C. L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27(1), 20–21 (2001).
Brunkow, M. E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27(1), 68–73 (2001).
Wildin, R. S. et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27(1), 18–20 (2001).
Reyahi, A. et al. Foxf2 is required for brain pericyte differentiation and development and maintenance of the blood–brain barrier. Dev. Cell 34(1), 19–32 (2015).
Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, the Stroke Genetics Network (SiGN), and the International Stroke Genetics Consortium (ISGC). Identification of additional risk loci for stroke and small vessel disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 15(7), 695–707 (2016).
French, C. R. et al. Mutation of FOXC1 and PITX2 induces cerebral small-vessel disease. J. Clin. Investig. 124(11), 4877–4881 (2014).
Lai, C. S. et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413(6855), 519–523 (2001).
Kume, T. et al. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93(6), 985–996 (1998).
De Val, S. et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135(6), 1053–1064 (2008).
Seo, S. et al. Forkhead box transcription factor FoxC1 preserves corneal transparency by regulating vascular growth. Proc. Natl. Acad. Sci. USA 109(6), 2015–2020 (2012).
Kume, T. et al. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 15(18), 2470–2482 (2001).
Topczewska, J. M. et al. The winged helix transcription factor Foxc1a is essential for somitogenesis in zebrafish. Genes Dev. 15(18), 2483–2493 (2001).
Lay, K., Kume, T. & Fuchs, E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc. Natl. Acad. Sci. USA 113(11), E1506–E1515 (2016).
Wang, L. et al. Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science 351(6273), 613–617 (2016).
Omatsu, Y. et al. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature 508(7497), 536–540 (2014).
Nishimura, D. Y. et al. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am. J. Hum. Genet. 68(2), 364–372 (2001).
Lehmann, O. J. et al. Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am. J. Hum. Genet. 67(5), 1129–1135 (2000).
Lehmann, O. J. et al. Ocular developmental abnormalities and glaucoma associated with interstitial 6p25 duplications and deletions. Investig. Ophthalmol. Vis. Sci. 43(6), 1843–1849 (2002).
Mears, A. J. et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld–Rieger anomaly. Am. J. Hum. Genet. 63(5), 1316–1328 (1998).
Nishimura, D. Y. et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet. 19(2), 140–147 (1998).
Gruneberg, H. Congenital hydrocephalus in the mouse, a case of spurious pleiotropism. J. Genet. 45, 1–21 (1943).
Skarie, J. M. & Link, B. A. FoxC1 is essential for vascular basement membrane integrity and hyaloid vessel morphogenesis. Investig. Ophthalmol. Vis. Sci. 50(11), 5026–5034 (2009).
Aldinger, K. A. et al. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy–Walker malformation. Nat. Genet 41(9), 1037–1042 (2009).
Muggerud, A. A. et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 12(1), R3 (2010).
Ray, P. S. et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res 70(10), 3870–3876 (2010).
Du, J. et al. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res. Treat. 131(1), 65–73 (2012).
Johnson, J. et al. FOXC1 identifies basal-like breast cancer in a hereditary breast cancer cohort. Oncotarget 7(46), 75729–75738 (2016).
Chanda, B. et al. A novel mechanistic spectrum underlies glaucoma-associated chromosome 6p25 copy number variation. Hum. Mol. Genet. 17(22), 3446–3458 (2008).
Avasarala, J. R., Jones, J. R. & Rogers, C. R. Forkhead box C1 gene variant causing glaucoma and small vessel angiopathy can mimic multiple sclerosis. Mult. Scler. Relat. Disord. 22, 157–160 (2018).
Poretti, A. et al. Joubert syndrome: Neuroimaging findings in 110 patients in correlation with cognitive function and genetic cause. J. Med. Genet. 54(8), 521–529 (2017).
Soliman, N. A. et al. Clinical characterization and NPHP1 mutations in nephronophthisis and associated ciliopathies: A single center experience. Saudi J. Kidney Dis. Transpl. 23(5), 1090–1098 (2012).
Stokman, M. F. et al. Clinical and genetic analyses of a Dutch cohort of 40 patients with a nephronophthisis-related ciliopathy. Pediatr. Nephrol. 33, 1701–1712 (2018).
Beales, P. L. et al. New criteria for improved diagnosis of Bardet–Biedl syndrome: Results of a population survey. J. Med. Genet. 36(6), 437–446 (1999).
Slavotinek, A. M. McKusick–Kaufman syndrome. In GeneReviews(R) (eds Adam, M. P. et al.) (University of Washington, 1993).
Keppler-Noreuil, K. M. et al. Brain tissue- and region-specific abnormalities on volumetric MRI scans in 21 patients with Bardet–Biedl syndrome (BBS). BMC Med. Genet. 12, 101 (2011).
Riddle, R. D. et al. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75(7), 1401–1416 (1993).
Masuya, H. et al. A duplicated zone of polarizing activity in polydactylous mouse mutants. Genes Dev. 9(13), 1645–1653 (1995).
He, M., Agbu, S. & Anderson, K. V. Microtubule motors drive Hedgehog signaling in primary cilia. Trends Cell Biol. 27(2), 110–125 (2017).
Lee, R. T., Zhao, Z. & Ingham, P. W. Hedgehog signalling. Development 143(3), 367–372 (2016).
Ingham, P. W., Nakano, Y. & Seger, C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12(6), 393–406 (2011).
Kim, J., Kato, M. & Beachy, P. A. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 106(51), 21666–21671 (2009).
Sasman, A. et al. Generation of conditional alleles for Foxc1 and Foxc2 in mice. Genesis 50(10), 766–774 (2012).
Zarbalis, K. et al. Meningeal defects alter the tangential migration of cortical interneurons in Foxc1hith/hith mice. Neural Dev. 7, 2 (2012).
Haldipur, P. et al. Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth. Elife 3, e03962 (2014).
Mishra, S. et al. Cerebrovascular defects in Foxc1 mutants correlate with aberrant WNT and VEGF-A pathways downstream of retinoic acid from the meninges. Dev. Biol. 420(1), 148–165 (2016).
Siegenthaler, J. A. et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell 139(3), 597–609 (2009).
Anvarian, Z. et al. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15(4), 199–219 (2019).
Nachury, M. V. The molecular machines that traffic signaling receptors into and out of cilia. Curr. Opin. Cell Biol. 51, 124–131 (2018).
Carter, C. S. et al. Abnormal development of NG2+PDGFR-alpha+ neural progenitor cells leads to neonatal hydrocephalus in a ciliopathy mouse model. Nat. Med. 18(12), 1797–1804 (2012).
Mykytyn, K. et al. Bardet–Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA 101(23), 8664–8669 (2004).
Mykytyn, K. et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat. Genet. 28(2), 188–191 (2001).
Soriano, P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124(14), 2691–2700 (1997).
Morrison-Graham, K. et al. A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development 115(1), 133–142 (1992).
Stephenson, D. A. et al. Platelet-derived growth factor receptor alpha-subunit gene (Pdgfra) is deleted in the mouse patch (Ph) mutation. Proc. Natl. Acad. Sci. USA 88(1), 6–10 (1991).
Gruneberg, H. A. T. & Truslove, G. M. Two closely linked genes in the mouse. Genet Res. Camb. 1, 69–90 (1960).
Rozet, J. M. & Gerard, X. Understanding disease pleiotropy: From puzzle to solution. Sci. Transl. Med. 7(291), 291fs24 (2015).
Quisling, R. G., Barkovich, A. J. & Maria, B. L. Magnetic resonance imaging features and classification of central nervous system malformations in Joubert syndrome. J. Child Neurol. 14(10), 628–635 (1999) (discussion 669–72).
Kallakuri, S. et al. Endothelial cilia are essential for developmental vascular integrity in zebrafish. J. Am. Soc. Nephrol. 26(4), 864–875 (2015).
Alvarez, J. I. et al. The Hedgehog pathway promotes blood–brain barrier integrity and CNS immune quiescence. Science 334(6063), 1727–1731 (2011).
Yuan, K. et al. Activation of the Wnt/planar cell polarity pathway is required for pericyte recruitment during pulmonary angiogenesis. Am. J. Pathol. 185(1), 69–84 (2015).
Yao, Q. et al. Sonic hedgehog mediates a novel pathway of PDGF-BB-dependent vessel maturation. Blood 123(15), 2429–2437 (2014).
Portal, C. et al. Primary cilia deficiency in neural crest cells models anterior segment dysgenesis in mouse. Elife 8, e52423 (2019).
Seo, S. et al. Primary cilium in neural crest cells crucial for anterior segment development and corneal avascularity. Investig. Ophthalmol. Vis. Sci. 65(3), 30 (2024).
Almubarak, A. et al. Loss of Foxc1 and Foxc2 function in chondroprogenitor cells disrupts endochondral ossification. J. Biol. Chem. 297(3), 101020 (2021).
Kamentsky, L. et al. Improved structure, function and compatibility for Cell Profiler: Modular high-throughput image analysis software. Bioinformatics 27(8), 1179–1180 (2011).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9(7), 676–682 (2012).
Kume, T., Deng, K. & Hogan, B. L. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127(7), 1387–1395 (2000).
Andrae, J. et al. A role for PDGF-C/PDGFRα signaling in the formation of the meningeal basement membranes surrounding the cerebral cortex. Biol. Open 5(4), 461–474 (2016).
Acknowledgements
We are grateful to the patients who participated in this study. We thank Drs. Sudipto Roy (National University of Singapore), Michael Walter (University of Alberta), Peter Carlsson (University of Gothenburg), and Valerie Wallace (University of Toronto) for critically reviewing earlier versions of the manuscript. Funding was provided by the Canadian Institutes of Health Research (CIHR) (MOP-133658) and Women and Children's Health Research Institute (to OJL), Natural Sciences and Engineering Research Council (to AJW and FBB) and Alberta Innovates Health Solutions (to IMM).
Funding
This study was funded by Alberta Innovates—Health Solutions, National Sciences and Engineering Research Council of Canada (No. NSERC RGPIN-2016-04682), Canadian Institutes of Health Research (No. MOP-133658), Women and Children’s Health Research Institute (No. 3149).
Author information
Authors and Affiliations
Contributions
OJL and SH conceived the study. SH performed and analyzed the majority of the experiments. PC, SVB, CRF and FBB contributed to specific experiments and data analysis. TK and AJW were involved in study design and supervision. IMM provided additional funding for the study, and with JRA and RCR contributed patient phenotypic data and reagents. SH and OJL co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Havrylov, S., Chrystal, P., van Baarle, S. et al. Pleiotropy in FOXC1-attributable phenotypes involves altered ciliation and cilia-dependent signaling. Sci Rep 14, 20278 (2024). https://doi.org/10.1038/s41598-024-71159-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71159-y