Abstract
Currently, surgical resection remains the primary approach for treating oral squamous cell carcinoma (OSCC), with limited options for effective drug therapy. Cardamonin, a principal chalcone compound derived from plants of the Zingiberaceae family, has garnered attention for its potential to suppress the onset and progression of various malignancies encompassing breast cancer, hepatocellular carcinoma, and ovarian cancers. Nevertheless, the involvement of cardamonin in the treatment of OSCC and its underlying mechanisms are yet to be elucidated. This research explored the possible target of cardamonin in treating OSCC via network pharmacological analysis. Subsequently, this research investigated the impact of cardamonin on OSCC cells via in vitro experiments, revealing its capacity to impede the migration, proliferation, and invasion of OSCC cells. Additionally, western blotting analysis demonstrated that cardamonin facilitates apoptosis by regulating the PI3K/AKT pathway. The findings suggest that MMP9 and the PI3K/AKT signaling pathway may serve as the target and pathway of cardamonin in treating OSCC. To summarize, the research findings suggest that cardamonin may facilitate apoptosis in OSCC cells by inhibition of PI3K/AKT pathway activation. These outcomes offer a theoretical basis for the utilization of cardamonin as a natural drug for treating OSCC.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) stands as the predominant type of head and neck tumors, comprising around 90% of oral malignant tumors1. Key contributors to its onset include tobacco, alcohol, and human papillomavirus (HPV) infections. Presently, surgical resection remains the primary treatment modality for OSCC, often complemented by postoperative chemoradiotherapy2. However, the 5-year survival rate remains dismally below 50%, exhibiting minimal improvement3.
In recent years, Chinese herbs have emerged as pivotal players in tumor treatment, serving as integral adjuncts to conventional therapies. Notably, certain chemotherapeutic agents, such as paclitaxel, vincristine4, and vinblastine5, derived from natural plant extracts, are extensively utilized in clinical settings. Cardamonin, a primary bioactive compound extracted from the ginger plant Myristica fragrans, exhibits a potent inhibitory effect on NF-kB6, STAT3, mTOR7, and Wnt/β-catenin signaling pathways8, all of which are widely involved in pro-inflammatory and pro-tumorigenic pathways. Consequently, cardamonin has been documented to exhibit antitumor9, anti-inflammatory10, and analgesic11 impacts, alongside immunomodulatory properties and enhancement of chemotherapy response rates. Increased research has showcased the capability of cardamonin to promote tumor cell apoptosis and suppress cell migration and invasion across several cancer types, encompassing breast cancer12, hepatocellular carcinoma13, colorectal carcinoma14, and oesophageal carcinoma15, thereby inhibiting tumor growth and metastasis. Nevertheless, research on the role of cardamonin in OSCC remains limited, with its underlying mechanism remaining elusive. Considering the simple surgical approach for OSCC and the potential side effects associated with conventional drug therapies, there is an urgent need to explore natural chemotherapy agents.
The inception of network pharmacology as a concept can be attributed to Andrew L. Hopkins in 2007, a period that coincided with significant strides in bioinformatics and systems biology16. This approach is grounded in systems biology, aiming to construct a multilevel network encompassing genetic-disease and disease-disease interactions. Its primary objective is to elucidate the relationships between active medicinal components and diseases17. In addition, molecular docking, a computer-aided drug design method, enables the prediction of potential ligand–protein bindings by analyzing the structural characteristics of substances18. This study advocates for an integrated approach that integrates molecular docking, network pharmacology, and experimental methods to analyze potential targets. PI3K/Akt pathway transmits extracellular signals into cells to regulate various pathologic processes, such as cell metabolism, proliferation, It plays a key role in migration, apoptosis, vesicle transport, canceration and chemotherapy resistance19,20. The interaction between PI3K and Akt. It is the key core of the function of this pathway21. MMPs are a family of zinc-dependent, calcium-regulated endogenous proteases. It is usually produced in its precursor form (pro-MMPs), is activated by other enzymes or free radicals through cysteine switching mechanism22, and participates in extracellular matrix remodeling under normal physiological and pathological conditions23, promotes the transformation of various extracellular matrix proteins, plays an important role in the process of tissue remodeling, and leads to structural changes in the cellular and tissue environment. MMP-9, also known as gelatinase B, is often considered to be consistent with MMP-2 as a marker of tumor progression and is closely associated with tumor invasion and metastasis. MMP-9 degrades extracellular matrix, activates interleukin-1β, and dissolves and cuts several chemokines24. The comprehensive scheme of the research is depicted in Fig. 1.
Materials and methods
Acquisition of cardamonin targets
The SMILES structural formula of cardamonin was retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/)25 and subsequently imported into the Swiss Target Prediction database (http://www.swisstargetprediction.ch/)26 to identify potential targets of cardamonin.
Screening potential targets of cardamonin for the treatment of oral squamous cell carcinoma
Using "Oral squamous cell carcinoma" as the keyword in both the Gene Card (https://www.genecards.org/)27 and OMIM database (https://www.omim.org/), disease-related targets were obtained. Duplicate targets identified in both databases were subsequently removed to isolate targets specifically related to OSCC. Following this, all identified targets associated with OSCC were imported into the online mapping tool (http://www.bioinformatics.com.cn/static/others/jvenn/example.html) to generate Venn diagram, thereby identifying intersectant targets between drugs and diseases.
Establishment of PPI network and target screening
The intersecting targets of drugs and diseases were imported into the String database (https://string-db.org/cgi/input.pl), with "Homo Sapiens" specified as the species for analysis. The confidence parameter was set to 0.4, and unassociated target proteins were removed. Subsequently, the Cytoscape 3.9.1 software was utilized to generate PPI network diagrams. Cytoscape is a sophisticated network analysis platform renowned for its robust data integration and visualization functions28. Core targets were obtained using the Centiscape2.2 plug-in acquisition.
GO and KEGG pathway enrichment analysis
The identified possible targets underwent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) functional enrichment analyses utilizing the DAVID database. The species was set as "Homo sapiens," with a statistical significance threshold set at P < 0.01. Subsequently, GO function and KEGG pathway enrichment analysis, along with visualization of the findings, were conducted utilizing the Microbiotics online tool (http://www.bioinformatics.com.cn/).
Molecular docking and visualization analysis
Molecular docking is a method of predicting the preferred direction of a molecule (ligand) when it binds to another molecule (receptor, such as RNA or enzyme). In this study, we adopted a semi-flexible docking method to form a stable composite. This process is critical for explaining mechanisms of action or screening lead compounds, and thus becomes one of the fundamental approaches for structure-based drug design. AutoDock Vina 1.1.2 software was used to combine cardamonin (PubChem CID: 641785) with proteins MMP9 (PDB ID: 1L6J), EGFR (PDB ID: 6TFV), ESR1 (PDB ID: 1HCQ), HSP90AB1 (PDB ID: 6N8Y), PPARG (PDB ID: 1PRG) and PTGS2 (PDB ID: 5F19) were used for molecular docking. 3D models of the proteins can be downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/pdb). The protein was pre-treated with PyMol 2.4 (removing water molecules and excess ligands, adding hydrogen atoms). AutoDock Tools 1.5.6 is used to generate PDBQT files for docking simulation. Set the docking result to output the 10 best docking positions. The docking conformation with the lowest binding energy and the highest clustering frequency is considered to be the most potential binding mode between ligands and proteins. Finally, we used Pymol 2.0 software to visualize the docking results. In this way, we can visually observe the binding of the ligand to the receptor and further analyze the stability and interaction of the complex.
Chemicals and reagents
Cardamonin (purity > 95%) was procured from Solarbio (Beijing, China) and dissolved in DMSO (Dimethyl sulfoxide) (Beijing, Solarbio) to reach a concentration of 200 µmol/l. In the process of preparing the protein sample, Protein Phosphatase Inhibitor (P1260-1 ml, Beijing Solarbio) and PMSF (Phenylmethylsulfonyl fluoride) (P0100-1 ml, Beijing, Solarbio) were included. The following antibodies were utilized: PI3K (AF6241), p-PI3K (AF3241) (both 1:1000, Affinity), AKT (ET1609-47), p-AKT (ET1607-73) (both 1:500, HuaBio), Bax (ab32503), Caspase-3 (ab184787) and Bcl-2 (ab196495) (all 1:2000, Abcam). Additionally, 740 Y-P (HY-P0175), a potent, cell-permeable PI3K activator, was acquired from MedChemExpress (MCE, USA).
Cell culture
The OSCC cell lines HSC-3 and CAL-27, identified by Short Tandem Repeat(STR) analysis, along with the normal oral mucosal epithelial cell line HOK, were cultured in Dulbecco's Modified Eagle Medium(DMEM) medium encompassing 1% penicillin and streptomycin and 10% Fetal Bovine Serum(FBS). The culturing procedure was carried out in a humid environment in an incubator with 5% CO2 at 37 °C. The selection of CAL-27 and HSC-3 cell lines was based on our specific research content. Both CAL-27 and HSC-3 are tongue squamous cell lines, and tongue cancer has the highest metastasis rate among oral and maxillofacial cancers. Most of our research content is about migration and invasion, and MMP9, a molecule we screened, is also an important molecule in the EMT process, so we chose the above two cell lines in this study.
Cell counting kit‑8 (CCK‑8) assay
The cell density was adjusted to 5 × 103 cells per well, and the cells were seeded into 96-well plates. After an attachment period of 8 h, HSC-3, and CAL-27 cells underwent treatment with varying cardamonin concentrations (0, DMSO, 10, 20, 40, 80, and 160 µmol/L), with DMSO concentration representing the concentration added during cardamonin dilution. A control group with only DMSO was included to account for its potential effect on the cells. Each group was set up with three replicate wells. Following incubation for 24 h, 10 µL of CCK-8 reagent was introduced to all wells, and the optical density (OD) value was measured at 450 nm utilizing an enzyme marker after a 2 h incubation period. The 50% inhibitory concentration(IC50)values for cardamonin concentrations of 0, 10, 20, and 40 µmol/L were selected for further investigation based on their respective values at 24 h.
Cell grouping and drug delivery methods
HSC-3 and CAL-27 cells underwent treatment with cardamonin (0, 10, 20, and 40 µmol/L), respectively.
Clone formation assay
The cell density was adjusted to 1 × 103 cells/well and seeded into 6-well plates. After cell attachment for 8 h, the cells underwent treatment as described previously, with each group having three replicate wells. Following 24 h of treatment with cardamonin, the complete medium was changed, and the cells underwent incubation for an additional 12 days. Subsequently, the cells underwent fixation utilizing 4% paraformaldehyde and staining using 0.1% crystal violet. The quantification of colony numbers was conducted utilizing ImageJ software.
Wound healing assay
CAL-27 and HSC-3 cells were seeded into 6-well plates at a density of 3 × 105 cells per well, with 2 ml of 10% FBS medium. The plates were then kept in an incubator with 5% CO2 and a constant temperature of 37 °C for cell culture. Once the cell density reached 100%, use the yellow tip to draw a straight line horizontally across the diameter of the six-well plate.. Caution was taken to ensure uniformity in drawing the line to maintain consistent well widths. Immediately after drawing the line, photos were captured using an inverted microscope, and the photo position of each well was recorded. Subsequently, 2 ml of DMEM culture medium without FBS was introduced into each well, and the cells were treated with cardamonin according to the same treatments and groupings as before. After 24 h of cardamonin treatment, the cell scratch healing rate was determined at the same position where the line was drawn initially. The following formula was utilized to calculate the cell scratch healing rate: Cell scratch healing rate = [(0 h scratch area- 24 h scratch area)/0 h scratch area] × 100%.
Transwell assays
Before the experiment, the matrix gel was diluted at a ratio of 1:8 with DMEM culture medium without FBS and set aside. The diluted matrix gel was introduced vertically into the inner lateral membrane of the chambers. HSC-3 and CAL-27 were digested and resuspended with a double-free medium containing different concentrations of cardamonin, treated and grouped as before. The cells were counted and inoculated on the inner membrane of the chamber at a density of 5 × 104 cells/well, with 200ul 10% FBS medium. Different concentrations of cardamonin were added to the corresponding well plates below the chamber, and the cells were treated and grouped as before. After 24 h, the medium in the 24-well plate and the chamber was removed, and the cells were fixed by adding 4% paraformaldehyde solution. Crystal violet staining solution was then introduced to stain the cells. The plate underwent washing with PBS three times, and the inside of the chamber was gently wiped with a cotton swab to eliminate excess dye. Subsequently, the plate was air-dried, images were captured, and the number of cells in each field of view was quantified.
Western blotting analysis
The cell lysates were prepared on ice utilizing RIPA buffer, PMSF and Protein Phosphatase Inhibitor (All-in-one, 100x) (Solarbio, Beijing, China), after which centrifugation was executed to acquire the supernatant. Subsequently, its protein concentration was assessed utilizing a BCA kit (Solarbio). Next, the proteins underwent denaturation by heating at 100 °C. Equivalent quantities of protein were isolated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, with a subsequent transfer onto polyvinylidene difluoride membranes. The membranes then underwent blocking with 5% milk in TBS-T at room temperature for 2 h. Subsequent to blocking, the membranes underwent overnight exposure to the primary antibody. Then, the membranes were further exposed to secondary antibodies at room temperature. Visualization of the protein bands was conducted utilizing an ECL kit, and densitometric analysis was executed utilizing ImageJ software.
Flow cytometry
CAL-27 and HSC-3 cells were inoculated into 6 cm diameter dishes with 3 mL of 10% FBS medium/well. The dishes were then placed in an incubator with 5% CO2 at a constant temperature of 37 °C for cell culture. Once the cell density reached 50–70%, the cells were treated with cardamonin as previously described. Following 24 h of cardamonin treatment, the cells underwent digestion with EDTA-free trypsin, followed by centrifugation to discard the supernatant. After this, cells underwent washing thrice with PBS solution and then were resuspended in 500 µL PBS solution. The cells were then stained following the procedure provided in the Annexin V-FITC/PI double-labeling staining kit. Subsequently, the stained cells were examined utilizing a flow cytometer, and the sums of early and late apoptosis were calculated for each group.
Statistical analysis
Data were examined utilizing GraphPad Prism v9.0 statistical software. Each experiment was replicated three times. The data are presented as the mean ± standard deviation (SD). Variations between two or more groups were assessed utilizing the Student’s t-test or one-way analysis of variance. P < 0.05 was deemed as a statistically significant value.
Results
Targets of cardamonin in oral squamous cell carcinoma
Cardamonin, a chemical substance typically extracted from plants within the Zingiberaceae (ginger) family is depicted by its chemical structural formula in Fig. 2a. To elucidate potential targets of cardamonin in OSCC, the Canonical SMILES number of cardamonin was retrieved from the PubChem database. Subsequently, 100 targets associated with the activity of cardamonin were identified through the Swiss Target Prediction database.
PPI networks and key targets. (a) Chemical structure of cardamonin. (b) Venn diagram of intersecting targets of cardamonin and OSCC, with 72 intersecting targets. (c) PPI network of cardamonin for OSCC. (d) Key target genes of cardamonin against OSCC. The redder and larger the node, the more significant it is in the network.
To further narrow down potential targets relevant to OSCC, 6729 targets related to OSCC were screened using the GeneCards database. From this screening, 3089 genes associated with OSCC were identified based on association scores exceeding the median value of 14.45.
Additionally, 429 target genes associated with OSCC were obtained from OMIM. By merging and eliminating duplicate data, 3351 OSCC-related target genes were compiled.
By intersecting these datasets, 72 potential targets of cardamonin in OSCC were identified using the Wayne online tool (Fig. 2b).
PPI network analysis and key target screening
The intersecting targets of cardamonin and OSCC were uploaded to the String platform, with the species set as "Homo sapiens", and unconnected nodes in the network were hidden. Default parameters were applied to acquire protein interaction data and construct the PPI network (Fig. 2c), which comprised 72 nodes interacting through 417 edges.
Subsequently, the protein interaction data were imported into Cytoscape 3.7.1 software. Topological analysis was conducted using the Centiscape2.2 plug-in, resulting in the identification of 14 core targets based on Degree ≥ 11.58333, Betweenness ≥ 73.94444, and Closeness ≥ 0.007040748 criteria. The screening yielded 14 nodes and 67 edges, which were then visualized and analyzed (Fig. 2d).
The analysis revealed that ESR1, EGFR, CXCR4, MMP9, HSP90AB1, PTGS2, PPARG, APP, FLT1, MCL1, PRKCA, PTK2, TERT, and HSPA8 are the core target genes of cardamonin in its action against OSCC.
GO enrichment analysis
The 72 potential targets of cardamonin acting on OSCC underwent GO enrichment analysis, which encompassed three parts: biological process (BP), cellular component (CC), and molecular function (MF)29. The GO enrichment analysis generated 288 GO items, including 172 BPs, 40 CCs, and 76 MFs.
After excluding the enrichment results with P > 0.05 and sorting them according to the count value, the top 10 items of each module were visualized using the microbiology letter platform (Fig. 4a). The BP category mainly involved negative regulation of transcription from RNA polymerase II promoter, negative regulation of apoptotic signaling pathway, positive regulation of cell proliferation, etc. The CC category mainly involved the cytosol, plasma membrane, etc. The MF category primarily involved ATP binding, enzyme binding, protein binding, etc., indicating the importance of proteins in disease.
KEGG enrichment analysis
Pathways with P < 0.05 were screened according to the enrichment results, resulting in 59 KEGG pathways that were enriched and sorted as per the COUNT value. The top 20 pathways were chosen to create visualized bubble diagrams using the microbiology letter platform (Fig. 4b). The results showed that the relevant signaling pathways of cardamonin acting on OSCC were the PI3K-Akt and MAPK pathways, among others. Notably, the PI3K/AKT pathway exhibited a higher degree of enrichment.
The PI3K-Akt pathway is a key signaling pathway regulating cell proliferation and motility, thereby contributing to cell survival. Its activation can contribute to the aberrant proliferation of OSCC cells.
Molecular docking
Molecular docking has been widely used to study possible binding patterns between molecules. In this study, molecular docking technology was used to explore the optimal binding mode of cardamonin to six proteins (Fig. 3). The docking results listed the binding energy, interaction force and bond length information for molecular docking (Table 1). The binding energy of cardamonin with MMP9, EGFR, ESR1, HSP90AB1, PPARG and PTGS2 reached − 8.5 kcal/mol, − 7.5 kcal/mol, − 6.2 kcal/mol, − 8.3 kcal/mol, respectively. − 7.7 kcal/mol and − 8.1 kcal/mol are bonded by hydrogen bonding, hydrophobic interactions and π-stacking. Among them, MMP9 has the lowest binding energy and the best binding effect.
Expression level analysis of core target genes in head and neck squamous carcinoma
To explore the expression level of the core target gene MMP9 in relation to the development of head and neck squamous carcinoma (HNSC), gene expression data were retrieved from The Cancer Genome Atlas (TCGA) database. The expression data of the target gene MMP9 were assessed in individuals with HNSC and compared with the normal population.
The outcomes highlighted that the expression of MMP9 was increased in HNSC patients relative to the normal population, and this difference was observed to be statistically significant (Fig. 4c).
Common target gene enrichment analysis and validation of important target genes. (a) Functional enrichment analysis encompassing BP, CC, and MF categories. (b) KEGG pathway enrichment analysis. Dot size and color represent the no. of genes and corrected P-value, respectively. (c) Data from the UALCAN database show that MMP9 exhibits enhanced expression in OSCC.
Cardamonin inhibited oral squamous cell carcinoma cell proliferation in a concentration-dependent manner
To examine the impact of cardamonin on the activity of OSCC cell lines CAL-27 and HSC-3, along with human normal keratinocytes (HOK), cells underwent treatment with increasing concentrations of cardamonin (0, 10, 20, 40, 80, 160 μmol/L) for 24 h. Subsequently, the IC50 values of the drug required to inhibit 50% of cell growth (IC50) were determined utilizing the CCK-8 assay. The findings highlighted that the IC50 values for CAL-27 and HSC3 cells were 41.28 μmol/L and 41.24 μmol/L, respectively. This indicates that the activity of OSCC cells decreased in a concentration-dependent manner with increasing concentrations of cardamonin (Fig. 5a–c). However, at concentrations below 80 μmol/L of cardamonin in the treatment of human HOK cells, cardamonin did not exhibit a considerable inhibitory impact on the activity of HOK cells. This suggests that low concentrations of cardamonin do not have significant toxicity to normal oral epithelial cells. Conversely, cardamonin demonstrated a considerable inhibitory impact on the proliferation of OSCC cells. This finding supports the safety of using cardamonin for the treatment of OSCC. To further elucidate the impact of cardamonin on the proliferative capacity of OSCC cells, a clone formation assay was utilized to examine the clone formation ability of CAL-27 and HSC-3 cells subjected to treatment with various concentrations of cardamonin. The findings highlighted that cardamonin displayed a concentration-dependent inhibitory impact on the proliferative capacity of OSCC cells. Notably, nearly complete inhibition of cancer cell proliferation was observed when treated with 40 μmol/L of cardamonin (Fig. 5d). These results underscore the concentration-dependent inhibition of OSCC cell proliferation by cardamonin.
Effect of cardamonin on OSCC cell activity. Cells underwent treatment with various concentrations (0, 10, 20, 40, 80, 160 µmol/L) for 24 h, and the cytotoxic impact of cardamonin on CAL27 cells (a) and HSC3 cells (b) was detected utilizing the CCK-8 assay. (c) The impact of cardamonin on the activity of three types of cells, namely, CAL-27, HSC-3, and HOK, was detected using the CCK-8 assay. (d) Cells underwent treatment with various concentrations (0, 10, 20, and 40 µmol/L) for 12 d. The clone formation ability of cardamonin on CAL27 and HSC3 cells was examined by cell clone formation assay. The data were expressed as mean ± SD (n = 3) and compared with the control group, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cardamonin inhibited oral squamous cell carcinoma cell migration and invasion in a concentration-dependent manner
To investigate the impact of cardamonin on the migratory and invasive capabilities of OSCC cells, scratch assays were carried out to determine the migratory capacity of OSCC cells following treatment with various concentrations of cardamonin (0, 10, 20, and 40 μmol/L). The results demonstrated a significant inhibitory effect on the migration of CAL-27 and HSC-3 cells treated with cardamonin, with the inhibitory effect being concentration-dependent (Fig. 6a). Additionally, transwell migration and invasion assays were executed, revealing a reduction in both migratory and invasive abilities of CAL-27 and HSC-3 cells following treatment with cardamonin (Fig. 6b). MMP9 emerges as the primary binding target for cardamonin in OSCC cells, functioning as a pivotal regulator in the epithelial-mesenchymal transition (EMT) process. EMT, known for promoting cell migration and invasion30, implies that cardamonin might impede these processes by binding to MMP9 and inhibiting EMT. To substantiate this hypothesis, this study assessed the expression of molecules related to EMT. The findings revealed that cardamonin upregulated the epithelial marker protein E-cadherin while reducing the levels of mesenchymal proteins, including N-cadherin, vimentin, and snail. Additionally, the expression of MMP9 protein, a key target of cardamonin, was also downregulated (Fig. 6c,d). These findings strongly indicate that cardamonin exerts its inhibitory impact on OSCC cell migration and invasion by targeting MMP9 to suppress the occurrence of EMT.
Migration and invasion of CAL-27 and HSC-3 cell lines and expression of related proteins. (a) Cell migration of CAL-27 and HSC-3 cell lines at different concentrations of cardamonin experimental group compared with 0 concentration. (b) Cell invasive migration of CAL-27 and HSC-3 cell lines at different concentrations of cardamonin experimental group compared with 0 concentration. (c,d) The protein banding display and relative gray value analysis and relative protein expression of E-cadherin, N-cadherin, MMP9, Vimentin, and Snail in each group of CAL-27 and HSC-3 cell lines were assessed. The findings highlighted that the expression of epithelial-associated protein E -cadherin was enhanced, and the expression of mesenchymal-associated proteins N -cadherin, Vimentin, and Snail was decreased. The OSCC target protein MMP9 was significantly reduced by cardamonin treatment. The variation in expression was statistically significant. Data were expressed as mean, standard deviation (n ≥ 3), ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cardamonin-induced apoptosis via the PI3K/AKT pathway
Further investigation was conducted to ascertain whether cardamonin inhibited cancer cell proliferation by facilitating apoptosis in OSCC cells. The outcomes suggested a considerable elevation in the proportion of Annexin V+ cells in CAL-27 and HSC-3 cells upon treatment with cardamonin, as determined by Annexin-V/PI flow cytometry. Notably, the proportion of Annexin V+ cells increased with higher concentrations of cardamonin (Fig. 7a), indicating that cardamonin promotes apoptosis of OSCC cells in a concentration-dependent manner.
Cardamonin promoted apoptosis in CAL-27, HSC-3 cells via PI3K/AKT signaling pathway. (a) Apoptosis of CAL-27 and HSC-3 cell lines in different concentrations of cardamonin experimental groups compared with 0 concentration. (b) Protein band display and relative gray value analysis of PI3K, p-PI3K, AKT, p-AKT, and relative expression of proteins in CAL-27 and HSC-3 cell lines. (c) Protein band display and relative gray value analysis of p-PI3K, PI3K, p-AKT, and AKT in CAL-27 and HSC-3 cell lines in all groups were performed, wherein protein bands were displayed and analyzed for relative gray value and relative expression of proteins. The results showed enhanced expression of BAX and Cleaved-caspase3 and reduced expression of Bcl-2 and pro-caspases3. (d) Protein expression levels of PI3K, p-PI3K, AKT, p-AKT, and relative expression of proteins in the reverse experiments of CAL-27 and HSC-3 cell lines. Expression differences were statistically significant. Data are expressed as mean, standard deviation (n ≥ 3), ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Apoptosis represents an active and signaling-dependent process crucial for regulating cell death. Caspase-3 serves as the effector molecule responsible for executing apoptosis. Its activation marks a critical step that irreversibly commits the cell to undergo apoptosis31. Within the Bcl-2 family, two key molecules are notably significant: Bcl-2 and Bax. Western blot analysis highlighted that cardamonin treatment reduced the expression of apoptosis-related proteins in CAL-27 and HSC-3 cells. Furthermore, cardamonin treatment led to a reduction in pro-caspase-3 expression and an increase in cleaved-caspase-3 expression (Fig. 7c). Notably, these effects became more pronounced with increasing concentrations of cardamonin. In the previously discussed network pharmacology analysis, cardamonin was proposed to potentially induce apoptosis in OSCC cells by partially modulating the PI3K-AKT pathway. Abnormal activation of the PI3K/Akt pathway is known to promote the progression of OSCC by suppressing apoptosis, increasing drug resistance, facilitating angiogenesis, promoting metastasis, and inducing EMT32. Subsequently, further experiments were conducted to validate the hypothesis. Western blot analysis revealed that treatment with cardamonin contributed to a reduction in the protein expression levels of p-PI3K and p-AKT in CAL-27 and HSC-3 cells (P < 0.05) (Fig. 7b). These findings highlighted that cardamonin induced apoptosis in OSCC cells by partially suppressing the PI3K/AKT pathway.
Finally, a reverse experiment was performed by adding a PI3K activator. Western blotting analysis results indicated that the expression of p-AKT, p-PI3K, and apoptosis-associated proteins in the group treated with both cardamonin and the PI3K activator notably rebounded compared to the group treated with cardamonin alone (Fig. 7d).
Discussion
OSCC stands as the most common cancer affecting the oral and maxillofacial region33. Despite significant strides made in the diagnosis and treatment of OSCC, its mortality rate remains high, with the 5-year survival rate remaining below 50%34. Typically, OSCC is treated through a comprehensive treatment strategy tailored to the type, location, and stage of cancer, often prioritizing surgical intervention followed by adjuvant chemoradiotherapy35. Post-surgery, OSCC patients commonly experience compromised functions such as chewing, swallowing, and speaking, consequently diminishing their quality of life and significantly impacting their physical and mental well-being36.
Natural phytochemicals are increasingly recognized as valuable adjuncts in postoperative care and as prophylactic measures against recurrence owing to their favorable tolerability, safety profile, low toxicity, and antioxidant properties37. Resistance to chemotherapeutic agents significantly impacts the effectiveness of OSCC treatment, and natural plant-derived compounds offer promise in improving therapeutic outcomes while mitigating side effects, besides their inherent ability to impede tumor progression. The impact of herbal components on OSCC has been extensively investigated. Studies have revealed that herbal compounds like cryptotanshinone38, glaucoma oleocalyxin A39, and duchesnea40 possess the ability to impede the growth of OSCC via various mechanisms and signaling pathways. Cardamonin has demonstrated its ability to facilitate tumor cell apoptosis in several cancer types, including breast12, liver41, and colorectal cancers14. These outcomes suggest the potential of cardamonin as a therapeutic agent in oncology. Cardamonin, as a natural active ingredient, can target various links of tumor occurrence and development through multiple ways, showing great potential in the prevention and treatment of malignant tumors. However, it also has a series of defects such as poor water solubility, low bioavailability and strong irritation42. In recent years, the application of nanotechnology and hydrogels has effectively improved the bioavailability, and further studies on the efficacy and toxicity reduction of cardamonin should be strengthened to improve the bioavailability. Drug combination has always been the core drug principle in the field of anti-tumor. Cisplatin, as a first-line high-efficiency broad-spectrum anti-tumor drug, has relatively serious adverse reactions. Cardamonin can significantly reduce cisplatin induced kidney injury and synergically promote the anti-tumor effect of cisplatin43. However, there is limited research on the therapeutic effects of cardamonin, specifically on OSCC.
This research examined the key targets and potential mechanisms of cardamonin in treating OSCC using an integrated approach that combines molecular docking, network pharmacology, and in vitro experiments. Our findings suggest that cardamonin promotes apoptosis in OSCC cells by partially inhibiting the PI3K/AKT pathway. Through PPI network analysis, MMP9, EGFR, PTGS2, PRKCA, PPARG, and FLT1 were identified as the key targets involved in the treatment process of cardamonin on OSCC.
Matrix metalloproteinase 9 (MMP9) is an enzyme produced by cells and belongs to the matrix metalloproteinase family. It serves pivotal functions in both physiological and pathological processes, encompassing cell migration, tissue repair, and cell proliferation44. However, overexpression or enhanced activity of MMP9 has been linked to several inflammatory conditions and diseases, encompassing chronic obstructive pulmonary disease45, cardiovascular disease46, and tumor metastasis47. Given their relevance to the occurrence and metastasis of OSCC, these pivotal targets exhibited stronger binding affinity for cardamonin during molecular docking studies, suggesting its potential as an anti-OSCC agent.
To further corroborate the efficacy of cardamonin in addressing OSCC, OSCC cells were exposed to various concentrations of the compound. Subsequently, the IC50 values for different cell lines were determined after 24 h. It was observed that cardamonin exhibited concentration-dependent inhibition of OSCC cell proliferation, as evidenced by both the CCK-8 assay and clone formation assay.
Apoptosis denotes an orderly regulated mechanism of cell demise, transpiring under either physiological or pathological circumstances48. Dysregulated apoptosis stands as a primary contributor to human cancer development. The majority of anticancer therapies are designed to induce apoptosis in tumor cells, aiming to eradicate malignant cells49. Our flow cytometry analysis highlighted that cardamonin can enhance apoptosis in a concentration-dependent manner. The Bcl-2 family members and caspases play pivotal roles in regulating cell apoptosis50. Within the Bcl protein family, Bcl-2 exerts an anti-apoptotic influence by impeding the release of cytochrome c, whereas Bax exerts a pro-apoptotic influence by facilitating cytochrome c release. The balance between Bcl-2 and Bax governs the apoptotic status of cells51. Caspase-3 serves as a downstream enzyme performing the apoptosis process and stands as the foremost regulator of cell demise. Upon cleavage by proteases, the Caspase-3 zymogen undergoes activation, yielding cleaved Caspase-3, thereby amplifying the protease cascade and ultimately instigating apoptosis within the nucleus52. This research found that cardamonin inhibited the expression levels of Bcl-2 and pro-Caspase-3, while significantly promoting the expression levels of Bax and cleaved Caspase-3 in OSCC cells. These findings further corroborate the notion that cardamonin has the potential to promote apoptosis in OSCC cells.
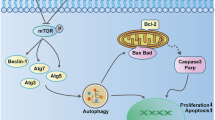
KEGG analysis revealed that cardamonin primarily regulates the RAS, PI3K/Akt, and MAPK signaling pathways in OSCC. Subsequent in vitro experiments demonstrated the inhibitory impact of cardamonin on PI3K activation. The PI3K-AKT pathway represents one of the most frequently dysregulated molecular pathways in human cancers, driving carcinogenesis by impeding normal apoptosis via the activation of various downstream effectors53. Previous studies underscore the involvement of the PI3K-Akt pathway in the development of multiple tumors. To further elucidate the inhibitory action of cardamonin on the PI3K/AKT pathway in OSCC, a significant reversal of the inhibitory impact of cardamonin was noted upon the addition of a PI3K agonist. This was accompanied by an elevation in the expression levels of p-PI3K and p-AKT, alongside a considerable elevation in the expression of apoptosis-related proteins encompassing BCL-2, pro-caspase 3, BAX, and cleaved-caspase 3 induced by the PI3K agonist. These outcomes highlight that cardamonin could facilitate OSCC cell apoptosis through the PI3K-Akt pathway. Furthermore, our findings revealed that cardamonin could impede tumor cell migration and invasion by inhibiting EMT. An abstract representation of the article is illustrated in Fig. 8.
Conclusion
This study highlights that cardamonin exhibits the potential to promote apoptosis in OSCC cells by suppressing the PI3K/AKT pathway activation. Furthermore, the results of this research lay a solid theoretical basis for the utilization of cardamonin as a natural drug in treating OSCC (Supplementary Information).
Data availability
All raw data are publicly available from corresponding databases. Processed data are available upon reasonable request from the corresponding author.
Change history
08 October 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-22476-3
References
Kina, S. et al. Higher overall survival rates of oral squamous cell carcinoma treated with metronomic neoadjuvant chemotherapy. Am. J. Cancer Res. 14(3), 1033–1051. https://doi.org/10.62347/EYNT8387 (2024).
Madera, M., Tirado Amador, L. & Leal, A. C. Therapeutic options in unresectable oral squamous cell carcinoma: A systematic review. Cancer Manag. Res. 13, 6705–6719. https://doi.org/10.2147/CMAR.S283204 (2021).
He, Y. et al. Lipid droplets-related Perilipin-3: Potential immune checkpoint and oncogene in oral squamous cell carcinoma. Cancer Immunol. Immunother. 73(5), 78. https://doi.org/10.1007/s00262-024-03659-9 (2024).
Mann, J. Natural products in cancer chemotherapy: Past, present and future. Nat. Rev. Cancer 2(2), 143–148. https://doi.org/10.1038/nrc723 (2002).
Zhu, H. et al. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biome. Pharmacother. 93, 1010–1017. https://doi.org/10.1016/j.biopha.2017.07.007 (2017).
Kubatka, P. et al. Cell plasticity modulation by flavonoids in resistant breast carcinoma targeting the nuclear factor kappa B signaling. Cancer Metastasis Rev. https://doi.org/10.1007/s10555-023-10134-x (2023).
Chen, H. et al. Cardamonin suppresses pro-tumor function of macrophages by decreasing M2 polarization on ovarian cancer cells via mTOR inhibition. Mol. Ther. Oncol. 26, 175–188. https://doi.org/10.1016/j.omto.2022.06.009 (2022).
Liu, Z., He, Z., Ai, X., Guo, T. & Feng, N. Cardamonin-loaded liposomal formulation for improving percutaneous penetration and follicular delivery for androgenetic alopecia. Drug Deliv. Transl. Res. https://doi.org/10.1007/s13346-024-01519-8 (2024).
Kim, Y. J., Kang, K. S., Choi, K. C. & Ko, H. Cardamonin induces autophagy and an antiproliferative effect through JNK activation in human colorectal carcinoma HCT116 cells. Bioorg. Med. Chem. Lett. 25(12), 2559–2564. https://doi.org/10.1016/j.bmcl.2015.04.054 (2015).
Chen, H., Shi, D., Niu, P., Zhu, Y. & Zhou, J. Anti-inflammatory effects of cardamonin in ovarian cancer cells are mediated via mTOR suppression. Planta Med. 84(16), 1183–1190. https://doi.org/10.1055/a-0626-7426 (2018).
Park, M. K. et al. Novel anti-nociceptive effects of cardamonin via blocking expression of cyclooxygenase-2 and transglutaminase-2. Pharmacol. Biochem. Behav. 118, 10–15. https://doi.org/10.1016/j.pbb.2013.12.019 (2014).
Jia, D. et al. Cardamonin reduces chemotherapy-enriched breast cancer stem-like cells in vitro and in vivo. Oncotarget 7(1), 771–785. https://doi.org/10.18632/oncotarget.5819 (2016).
Hossan, M. S. et al. Novel semi-synthetic Cu (II)-cardamonin complex exerts potent anticancer activity against triple-negative breast and pancreatic cancer cells via inhibition of the Akt signaling pathway. Molecules 26(8), 2166. https://doi.org/10.3390/molecules26082166 (2021).
Lu, T., Zheng, C. & Fan, Z. Cardamonin suppressed the migration, invasion, epithelial mesenchymal transition (EMT) and lung metastasis of colorectal cancer cells by down-regulating ADRB2 expression. Pharm. Biol. 60(1), 1011–1021. https://doi.org/10.1080/13880209.2022.2069823 (2022).
Wang, Z. et al. Cardamonin inhibits the progression of oesophageal cancer by inhibiting the PI3K/AKT signalling pathway. J. Cancer 12(12), 3597–3610. https://doi.org/10.7150/jca.55519 (2021).
Pei, H. et al. Network pharmacology and molecular docking analysis on the mechanism of Cordyceps militaris polysaccharide regulating immunity through TLR4/TNF-α pathwayss. J. Biochem. Mol. Toxicol. 37(6), e23345. https://doi.org/10.1002/jbt.23345 (2023).
Liu, S., Zhang, Q., Liu, W. & Huang, X. Research on the mechanisms of Shu Yu wan in the treatment of cervical cancer based on network pharmacology analyses and molecular docking technology. Nat. Prod. Rese. 37(4), 646–650. https://doi.org/10.1080/14786419.2022.2071884 (2023).
Wu, H. B., Xiao, Y. G., Chen, J. S. & Qiu, Z. K. The potential mechanism of Bupleurum against anxiety was predicted by network pharmacology study and molecular docking. Metab. Brain Dis. 37(5), 1609–1639. https://doi.org/10.1007/s11011-022-00970-1 (2022).
Navaei, Z. N. et al. PI3K/Akt signaling pathway as a critical regulator of cisplatin response in tumor cells. Oncol. Res. 29(4), 235–250 (2022).
Glaviano, A. et al. PI3K/Akt/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer. 22(1), 138 (2023).
Tewari, D. et al. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 80, 1–17 (2022).
Brkic, M. et al. Friends or foes: Matrix metalloproteinases and their multifaceted roles in neurodegenerative diseases. Mediators Inflamm. 2015, 620581. https://doi.org/10.1155/2015/620581.1 (2015).
Bonnans, C., Chou, J. & Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15(12), 786–801 (2014).
Opdenakker, G. et al. Gelatinase B functions as regulator and effector in leukocyte biology. J. Leukoc. Biol. 69(6), 851–859 (2001).
Kim, S., Chen, J. & Cheng, T. PubChem 2023 update. Nucleic Acids Res. 51(D1), D1373–D1380. https://doi.org/10.1093/nar/gkac956 (2023).
Gfeller, D., Grosdidier, A. & Wirth, M. L. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42(Web Server issue), W32-38. https://doi.org/10.1093/nar/gku293 (2014).
Rebhan, M., Chalifa-Caspi, V. & Prilusky, J. GeneCards: Integrating information about genes, proteins and diseases. Trends Genet. 13(4), 163. https://doi.org/10.1016/s0168-9525(97)01103-7 (1997).
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2(10), 2366–2382. https://doi.org/10.1038/nprot.2007.324 (2007).
Hopkins, A. L. Network pharmacology. Nat. Biotechnol. 25(10), 1110–1111. https://doi.org/10.1038/nbt1007-1110 (2007).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29(3), 212–226. https://doi.org/10.1016/j.tcb.2018.12.001 (2019).
Alzahrani, A. S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 59, 125–132. https://doi.org/10.1016/j.semcancer.2019.07.009 (2019).
Wang, J. et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 11(8), 682. https://doi.org/10.1038/s41419-020-02851-w (2020).
Ai, Y. et al. METTL3 intensifies the progress of oral squamous cell carcinoma via modulating the m6A amount of PRMT5 and PD-L1. J. Immunol. Res. 2021, 6149558. https://doi.org/10.1155/2021/6149558 (2021).
Wang, J., Du, X. X., Jiang, H. & Xie, J. X. Curcumin attenuates 6-hydroxydopamine-induced cytotoxicity by anti-oxidation and nuclear factor-kappa B modulation in MES23.5 cells. Biochem. Pharmacol. 78(2), 178–183. https://doi.org/10.1016/j.bcp.2009.03.031 (2009).
Sun, S. et al. MiR-302b suppresses tumor metastasis by targeting frizzled 6 in OSCC. J. Dent. Res. 100(7), 739–745. https://doi.org/10.1177/0022034520986551 (2021).
Wang, Z. et al. DRP1 inhibition-mediated mitochondrial elongation abolishes cancer stemness, enhances glutaminolysis, and drives ferroptosis in oral squamous cell carcinoma. Br. J. Cancer https://doi.org/10.1038/s41416-024-02670-2 (2024).
Ji, Y. et al. IL-1α facilitates GSH synthesis to counteract oxidative stress in oral squamous cell carcinoma under glucose-deprivation. Cancer Lett. 589, 216833. https://doi.org/10.1016/j.canlet.2024.216833 (2024).
Jiang, X. T., Qiu, Y. & Li, C. H. Cryptotanshinone inhibits oral squamous cell carcinoma through the autophagic pathway. Neoplasma 70(1), 114–122. https://doi.org/10.4149/neo_2023_220924N957 (2023).
Wang, X., He, M. J., Chen, X. J., Bai, Y. T. & Zhou, G. Glaucocalyxin A impairs tumor growth via amplification of the ATF4/CHOP/CHAC1 cascade in human oral squamous cell carcinoma. J. Ethnopharmacol. 290, 115100. https://doi.org/10.1016/j.jep.2022.115100 (2022).
Yang, W. E. et al. Duchesnea indica extract attenuates oral cancer cells metastatic potential through the inhibition of the matrix metalloproteinase-2 activity by down-regulating the MEK/ERK pathway. Phytomed. Int. J. Phytother. Phytopharmacol. 63, 152960. https://doi.org/10.1016/j.phymed.2019.152960 (2019).
Xu, Q. et al. Cardamonin reduces acetaminophen-induced acute liver injury in mice via activating autophagy and NFE2L2 signaling. Front. Pharmacol. 11, 601716. https://doi.org/10.3389/fphar.2020.601716 (2020).
Liu, Z., He, Z., Ai, X., Guo, T. & Feng, N. Cardamonin-loaded liposomal formulation for improving percutaneous penetration and follicular delivery for androgenetic alopecia. Drug Deliv. Transl. Res. 14(9), 2444–2460. https://doi.org/10.1007/s13346-024-01519-8 (2024).
El-Naga, R. N. Pre-treatment with cardamonin protects against cisplatin-induced nephrotoxicity in rats: Impact on NOX-1, inflammation and apoptosis. Toxicol. Appl. Pharmacol. 274(1), 87–95. https://doi.org/10.1016/j.taap.2013.10.031 (2014).
Hosseini, A., Kumar, S., Hedin, K. & Raeeszadeh-Sarmazdeh, M. Engineering minimal tissue inhibitors of metalloproteinase targeting MMPs via gene shuffling and yeast surface display. Protein Sci. 32(12), e4795. https://doi.org/10.1002/pro.4795 (2023).
Jiang, J. et al. CD146 deficiency aggravates chronic obstructive pulmonary disease via the increased production of S100A9 and MMP-9 in macrophages. Int. Immunopharmacol. 127, 111410. https://doi.org/10.1016/j.intimp.2023.111410 (2024).
Bhave, S. et al. Loss of bone morphogenetic protein-9 reduces survival and increases MMP activity after myocardial infarction. JACC Basic Transl. Sci. 8(10), 1318–1330. https://doi.org/10.1016/j.jacbts.2023.05.017 (2023).
Wu, S., Liu, L., Xu, H., Zhu, Q. & Tan, M. The involvement of MALAT1-ALKBH5 signaling axis into proliferation and metastasis of human papillomavirus-positive cervical cancer. Cancer Biol. Ther. 24(1), 2249174. https://doi.org/10.1080/15384047.2023.2249174 (2023).
Wong, R. S. Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30(1), 87. https://doi.org/10.1186/1756-9966-30-87 (2011).
Mohammad, R. M. et al. Broad targeting of resistance to apoptosis in cancer. Semin. Cancer Biol. 35(Suppl 0), S78–S103. https://doi.org/10.1016/j.semcancer.2015.03.001 (2015).
Herr, A. B. Evolution of an allosteric “off switch” in apoptotic caspases. J. Biol. Chem. 293(15), 5462–5463. https://doi.org/10.1074/jbc.H118.002379 (2018).
Ghobrial, I. M., Witzig, T. E. & Adjei, A. A. Targeting apoptosis pathways in cancer therapy. CA Cancer J. Clin. 55(3), 178–194. https://doi.org/10.3322/canjclin.55.3.178 (2005).
Crawford, E. D. & Wells, J. A. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 80, 1055–1087. https://doi.org/10.1146/annurev-biochem-061809-121639 (2011).
Wang, Y. et al. Erianin, the main active ingredient of Dendrobium chrysotoxum Lindl, inhibits precancerous lesions of gastric cancer (PLGC) through suppression of the HRAS-PI3K-AKT signaling pathway as revealed by network pharmacology and in vitro experimental verification. J. Ethnopharmacol. 279, 114399. https://doi.org/10.1016/j.jep.2021.114399 (2021).
Acknowledgements
Thanks to the Innovation Centre for Science and Technology of North Sichuan Medical College and the Institute of Hepatobiliary, Pancreatic and Intestinal Diseases of North Sichuan Medical College for their research platform and support. We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Funding
Funded Projects: Key Training Project of North Sichuan Medical College (CBY23-ZDA10), Cooperation Project of Nanchong University (22SXQT0356-Yu). The funder was not involved in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: Yuehan Wu, Yapei Wang, Xiaoxu Nan. Data curation: Yuehan Wu, Huan He. Formal analysis: Yuehan Wu. Methodology: Qiannan Hu, Yuqi Xie. Project administration: Ying Liu. Supervision: Ying Liu. Validation: Yuqi Xie. Visualization: Yuehan Wu. Writing—original draft: Yuehan Wu. Writing—review and editing: Yuehan Wu, Han Liu.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error, where a botanical classification of Myristica fragrans was incorrect. Modifications have been made to the Abstract and Results section. Full information regarding the corrections made can be found in the correction for this Article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Y., Wang, Y., Liu, H. et al. Mechanism of apoptosis in oral squamous cell carcinoma promoted by cardamonin through PI3K/AKT signaling pathway. Sci Rep 14, 20802 (2024). https://doi.org/10.1038/s41598-024-71817-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71817-1
Keywords
This article is cited by
-
Determination of prognostic parameters with impact on disease-free survival rate of patients treated for oral squamous cell carcinoma: a look at a few confounding variables
Odontology (2026)
-
Exploring anticancer potential of betanin in DMBA-induced oral squamous cell carcinoma: an in silico and experimental study
Naunyn-Schmiedeberg's Archives of Pharmacology (2025)
-
A Comprehensive Review on Oral Squamous Cell Carcinoma: Etiology, Current Therapeutic Strategies, Molecular Signalling Pathways, Herbal Therapeutics, and the Role of Targeted Vesicular Delivery Systems
BioNanoScience (2025)