Abstract
Mung bean (Vigna radiata L.), a widely cultivated legume, belongs to the Fabaceae family’s Papilionoideae subfamily. Although Phosphatidylethanolamine-binding protein (PEBP) genes have been identified in several plant species, their presence and function in mung bean remain largely unexplored. In this study, we identified seven VrPEBP genes from mung bean and classified them into four clades: FT, MFT, TFL and FT-like. Cis-regulatory element analysis revealed that VrPEBP genes may play a role in light, hormone, and stress responses. Quantitative real-time PCR (qRT-PCR) analysis indicated that VrPEBPs were constitutively expressed in various tissues. However, tissue-specific expression patterns were observed among VrPEBP genes. Under short-day (SD) conditions, VrFT1 and VrMFT1 exhibited significantly higher expression levels than under long-day (LD) conditions at 8 and 4 h, respectively. Conversely, VrTFL2 and VrTFL3 showed significantly higher expression levels under LD conditions compared to SD conditions at 8 and 12 h, respectively. The varied expression patterns of these genes under different photoperiod suggest their potential involvement in the photoperiodic regulation of flowering in mung bean. Additionally, phenotypic analysis of transgenic Arabidopsis plants overexpressing VrFT1 revealed higher expression levels under SD conditions and predicted its role in promoting flowering. These results provide valuable insights into the evolution and function of PEBP genes in mung bean and lay the foundation for further research on their regulatory mechanisms and potential applications in mung bean improvement.
Similar content being viewed by others
Introduction
Flowering is a crucial process for plant growth and development, as it determines the reproductive success and genetic diversity of plants. Flowering is regulated by various factors, such as photoperiod, temperature, and hormones1,2. Plants have adapted to various environmental conditions and have evolved different mechanisms to control their flowering time and pattern3. Understanding the molecular and physiological basis of flowering in plants can help improve crop yield and quality, as well as conserve plant biodiversity4. PEBP (phosphatidylethanolamine-binding protein) is a gene family that plays a vital role in regulating flowering time and pattern in plants. Different members of the PEBP family have different functions, such as promoting or inhibiting flowering, depending on the species and environmental conditions5. PEBPgenes are involved in various pathways, such as photoperiod, temperature, hormones, and genes that control flowering6. PEBPgenes are also highly diversified and evolved in different plant lineages, such as angiosperms and gymnosperms7. Studying the PEBP gene family can contribute to a better understanding of the molecular and physiological mechanisms of flowering in plants, thereby enhancing crop production and conservation efforts.
The PEBPgene family, present in various plant species, plays a crucial role in diverse biological processes, including the regulation of flowering time and the management of plant architecture1,2. This family encompasses three distinct categories of proteins: FT-Like Proteins: within Arabidopsis, the FT-like subfamily includes FT and its counterpart, TWIN SISTER OF FT (TSF), both of which facilitate the transition from vegetative to reproductive growth8. FT, an activator, migrates from leaves to the shoot apical meristem and interacts with the transcription factor FLOWERING LOCUS D (FD), thereby hastening the flowering process9. TSF, as a homologous gene to FT, accelerates early flowering in Arabidopsis and exhibits a redundant effect similar to FToverexpression10. TFL1-Like Proteins: Members of the TFL1-like subfamily, such as TFL1, Arabidopsis thaliana CENTRORADIALIS (ATC), and BROTHER OF FT (BFT), are known to delay the onset of flowering11,12. TFL1, through the regulation of meristem genes, inhibits flowering and governs plant architecture13. ATC, activated by short-day conditions, functions as a floral inhibitor and its overexpression extends the time to flowering14. BFT, closely related to FT and TFL1, causes delayed flowering when overexpressed in Arabidopsis, although its mutant form exhibits distinct phenotypic traits compared to the tfl1 mutant15. MFT Proteins: MOTHER OF FT AND TFL1 (MFT), an ancestral gene of both FT and TFL1, participates in seed germination and development. It is predominantly found in seeds and roots and is responsible for inducing seed dormancy16. Overexpression of AtMFTleads to early flowering, though not to the same extent as FT. Furthermore, MFT is predominantly expressed in seeds, primarily via the ABA and GA signaling pathways, and contributes to the regulation of seed germination17. The investigation of the PEBP gene family provides valuable insights into these intricate biological processes, with potential implications for crop development and conservation strategies.
The significance of flowering traits in crop production cannot be overstated. Among the core genes involved in the flowering process, the PEBP gene has garnered extensive attention and investigation across various plants. PEBP gene family is widely present in plants, and it has been identified in many other plants besides Arabidopsis. For example, PEBP gene family has been found in tomato, barley, grapevine, maize, apple, cotton, peanut and Dendrobium huoshanenseet al5,18,19,20,21,22,23,24.. These genes play important roles in the growth and development of plants, including regulation of flowering time, plant architecture and seed dormancy. Although these genes may differ in different plants, their functions are similar. For example, FT and TSF promote the transition from vegetative to reproductive growth, while TFL1 inhibits flowering and controls plant architecture. For instance, in rice (Oryza sativa), the Hd3a gene, which is an ortholog of the Arabidopsis FTgene, exhibits a fascinating movement from the leaf to the shoot apical meristem, ultimately triggering the flowering process25. Similarly, in soybean (Glycine max), the GmFT1a gene plays a pivotal role as a suppressor of flower formation. When exposed to alterations in photoperiod, soybean varieties sensitive to such changes exhibit delayed flowering. In contrast, the GmFT2a/5a gene acts as a promoter of flowering, showcasing an opposing function to GmFT1a26. In maize (Zea mays), the CENTRORADIALIS 8 (ZCN8) gene is of paramount importance for flower development. When subjected to a short-day treatment, ZCN8 demonstrates active expression in the shoot apical meristem, while its expression remains undetected under long-day conditions. This emphasizes the pivotal role of ZCN8in maize flower formation27.
Mung bean (Vigna radiataL.), a well-known pulse crop, belongs to the subfamily Papilionoideae of Fabaceae, which also includes soybeans, common beans, chickpeas, peanuts, and Medicago. Mung bean is widely cultivated across various regions, including Asia, Africa, South America, and Australia28,29,30. Mung beans are highly nutritious and provide significant health benefits. They are rich in plant-based protein, dietary fiber, vitamins (such as folate, vitamin C, and vitamin K), and minerals (including potassium, magnesium, and iron). These beans are also known for their low-fat content and potential antioxidant properties, which contribute to their positive impact on human health30. With the publication of the mung bean genome, a deeper understanding of its genetic makeup has been achieved31. This has facilitated the exploration and discovery of functional genes associated with various traits and processes. The availability of the genome sequence has greatly accelerated research on mung bean’s genetic improvement, disease resistance, and nutrient traits, leading to the development of high-yielding and more nutritious cultivars. Flowering plays a crucial role in reproduction and seed production. Research efforts aimed at unraveling the molecular mechanisms and regulatory factors involved in the flowering process have the potential to enhance crop yield and quality. To date, research on the PEBP gene family in mung beans has not been extensively conducted. As important regulators of flowering, the identification and analysis of members of the PEBP gene mung bean are crucial for deciphering the flowering time in mung bean.
In this study, we identified PEBP genes in mung bean, and comprehensively analyzed phylogenetic relationships, the chromosomal distributions, gene duplication, motif compositions, gene structures, and cis-acting elements of PEBP genes using bioinformatics methods. Their expression patterns in response to photoperiod were assessed using fluorescence quantitative PCR (qPCR) methods. Among these, key genes were functionally characterized in transgenic Arabidopsis to reveal its crucial role in photoperiod. Our findings lay the groundwork for further investigation into the functions of the PEBP families in mung bean. Additionally, this research will facilitate their utilization in the genetic improvement of crops.
Results
Identification and characteristics of the PEBP family members of mung bean
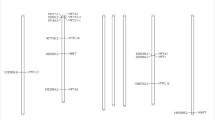
Seven PEBP family genes identified in mung beans (Table S1). The seven PEBP family genes identified in our study were distributed across chromosomes 1 (comprising three PEBP family genes), 3 (one gene), and 8 (three genes) (Fig. 1). Physicochemical analysis of VrPEBP family protein members revealed variations in the number of amino acids encoded by the seven VrPEBP genes, ranging from 154 (VrTFL3) to 556 (VrMFT1). The theoretical isoelectric point (pI) was found to display the lowest values in MFT proteins (e.g., 4.96 for VrMFT1) and FT-like (ranging from 4.68 for VrFT-like2 to 5.33 for VrFT-like1), while TFL proteins exhibited the highest values (e.g., 9.59 for VrTFL1, 9.07 for VrTFL2, and 9.60 for VrTFL3). The theoretical isoelectric point of the FT protein was determined to be 6.51 (VrFL1). The highest molecular weight was predicted by MFT proteins, with the lowest molecular weight guessed in TFL proteins (e.g., VrTFL3) (Table 1). The subcellular localization analysis of the seven VrPEBP genes indicated that predominant expression was observed in the cytoplasm (Table 1). This comprehensive examination underscores the significance of PEBP genes in mung bean and their distinct physicochemical characteristics.
Phylogenetic analysis of the PEBP genes in mung bean
In order to investigate the phylogenetic relationships within the PEBP gene family in mung bean, an extensive analysis was conducted on 163 PEBP members sourced from various plant species, including Arabidopsis thaliana, Phaseolus vulgaris, Glycine Max, Cicer arietinum, cultivated peanuts, Oryza Sativa, Zea mays, Sorghum bicolor, Vitis vinifera, and Medicago truncatula. . Based on the classification system of the PEBP family in A. thaliana, these genes were further categorized into distinct subclades, namely FT, TFL, MFT, and FT-like (Fig. 2). Among these subgroups, FT encompassed 79 members, TFL comprised 49 members, MFT contained 20 members, and FT-like consisted of 15 members. Notably, VrTFL1 was identified within the FT-like subclade, while VrTFL1, VrTFL2, and VrTFL3 were classified under the TFL branch. VrMFT1 was assigned to the MFT branch, and VrFT-like1 and VrFT-like2 were positioned within the FT-like branch. It is worth mentioning that mung bean PEBP proteins exhibited significant homology to protein within the corresponding clades of other plant species.
The constructed neighbor-joining phylogenetic tree was derived from 163 PEBP proteins obtained from various plant species, including Arabidopsis thaliana (At), Arachis hypogaea (Arahy), Cicer arietinum (Ca), Medicago truncatula (Mt), Phaseolus vulgaris (Pv), Glycine Max (Gm), Vitis vinifera (Vv), Oryza sativa (Os), Sorghum bicolor (Sb), Zea mays (Zm), and Vigna radiate (Vr). The 163 PEBPs were grouped into four distinct categories, each represented by a unique background color. Notably, PEBP proteins within the mung bean were highlighted with identifiable red circles.
Gene structure and conserved motifs analysis of VrPEBP genes
For an in-depth structure analysis of the PEBP gene family, we meticulously examined the structural aspects of the seven VrPEBP family members(Fig. 3a). In total, seven motifs were pinpointed in the VrPEBP proteins, designated as motifs 1 through 7, each spanning from 21 to 50 amino acids in length (Fig. 3b, Table S2, Figure S1). We observed that each PEBP family gene harbors 2–5 motifs. Specifically, the FT, TFL, and MFT subclades displayed five common motifs: motif 1, 2, 3, 4, and 5, while the FT-like subclade exhibited motifs 6 and 7. Notably, PEBP members within each branch exhibited variations in motif presence. The number of motifs in FT-type genes is consistently 5, while for FT-like genes, within the TFL subgroup, all except VrTFL3 contain 5 motifs. In the MFT subfamily, VrMFT1 contains 4 motifs.For instance, with the exception of VrTFL3, which solely possessed motifs 1–4, the other five PEBP genes in the TFL subclade displayed a consistent motif composition. In the MFT subclade, the two PEBP members exhibited a variation in conserved motifs, with one possessing 5 and the other 2. Furthermore, distinctive motif distributions were noted across different subclades. Motif 2 was exclusively present in subclades FT, TFL, and MFT, while Motif 6 and Motif 7 were exclusive to subclade FT-like. These disparities in motif distribution may underlie variations in PEBP gene structure, potentially leading to functional differentiation. To investigate the correlation between genome evolution and functional divergence, we conducted a thorough analysis of the VrPEBP gene structure. It was observed that, apart from VrMFT1, VrFT-like2, and VrFT-like1, the remaining five genes shared a common structure comprising four exons and three introns (Fig. 3d). In terms of phylogenetic relationships, closely related PEBP genes not only exhibited identical exon counts, but also displayed similar structural patterns. Through CDD analysis, we identified conserved PBP structural domains within each VrPEBP protein. Notably, these structural domains maintained consistent positions across different sequences (Fig. 3c). These findings underscored the high degree of conservation in the gene structure of VrPEBP family members. However, specific sequence structures were discernible within different PEBP gene subgroups, suggesting potential functional distinctions.
Phylogenetic tree, conserved motifs, gene structure and conserved domain of the PEBP protein in mung bean and Arabidopsis. (A) Phylogenetic tree of the PEBP protein sequences in mung bean and Arabidopsis species constructed by the Neighbor-Joining (NJ) method. (B) Conserved motifs in VrPEBPs using MEME-suite. Various colors represented different motifs. (C) A comparison of conserved domains between AtPEBPs and VrPEBPs was conducted, with the ruler at the bottom indicating sequence lengths. (D) The eight mung bean PEBP genes structures. The ruler at the bottom indicates length of the sequences.
Gene duplication and syntenic analysis of VrPEBP
In the study conducted on mung bean, an intraspecific collinearity analysis of PEBP genes revealed the absence of collinear genes (Fig. 4a). Subsequently, an extended analysis was performed to evaluate interspecific collinearity relationships, providing insights into the evolutionary connections of mung bean with other plants.. The findings revealed diverse collinearity relationships, with 44, 8 and 11 pairs of collinearity connections identified between VrPEBP gene family members and other legumes, dicotyledonous plants, and monocotyledonous plants, respectively (Fig. 4b). Additionally, there were 8, 8, 6, 5 and 17 gene pairs exhibiting collinearity between mung bean and other legumes, such as soybean, alfalfa, chickpea, common bean and cultivated peanuts. Furthermore, 6 and 2 gene pairs were identified between mung bean and the dicotyledonous plants grape and Arabidopsis, respectively, while 4, 4 and 3 gene pairs were found between mung bean and the monocotyledonous plants rice, sorghum and maize. The collinearity analysis results indicated a closer relationship between mung bean and legume plants in terms of PEBP genes compared to dicotyledonous and monocotyledonous plants, with the closest relationship observed with cultivated peanuts. Orthologous gene pairs were also explored, revealing variations in synteny relationships among VrPEBP genes and different species (in Table S3). For instance, VrTFL2 in the TFL subclade displayed the highest number of orthologous gene pairs (28 pairs) with other species, followed by VrTFL3 in the TFL subclade, which had 11 pairs of orthologous gene pairs. In contrast, VrFT-like2 in the FL-like subclade exhibited the fewest number of orthologous gene pairs (3 pairs) with other species. All VrPEBPs exhibited synteny relationships with cultivated peanuts, while mung bean and monocotyledonous plants shared synteny only in the TFL subclade. These findings suggest that distinct VrPEBP genes evolved gradually through the replication of synteny modules during the course of evolution.
Syntenic analysis of PEBP genes between mung bean and other plants. (A) Syntenic relationships of PEBP genes with in mung bean. The putative homologous PEBP genes are linked by gray colored lines. (B) Syntenic analysis of PEBP genes between mung bean and other plants, including Glycine Max (Gm), Cicer arietinum (Ca), Medicago truncatula (Mt), Phaseolus vulgaris (Pv), Arabidopsis thaliana (At), Arachis hypogaea (Arahy), Vitis vinifera (Vv), Oryza sativa (Os), Sorghum bicolor (Sb) and Zea mays (Zm). Gray lines in the background indicated the collinear blocks with in mung bean and other plant genomes, while the syntenic PEBP gene pairs are linked with red lines.
To assess the evolutionary pressures acting on the VrPEBP genes, the nonsynonymous substitution rate (Ka), synonymous substitution rate (Ks), and their ratio (Ka/Ks) were computed for homologous gene pairs (Table S4). Notably, the Ka/Ks ratios for 53 of the collinear gene pairs between mung bean and other species were all less than 1, indicating that VrPEBP genes primarily underwent purifying selection during their evolutionary journey.
Promoter analysis of VrPEBPs
In this study, multiple cis-acting promoter elements were observed in the VrPEBP gene family, and they play a crucial role in regulating gene expression and function. These cis-acting elements fall into three primary categories (Table S5; Fig. 5): light-responsive elements, stress-responsive elements, and hormone-responsive elements. Specifically, the cis-acting promoter elements of PEBP gene family can be categorized into three groups, each with cis-acting elements number: 83 light-responsive elements, 29 stress-responsive elements, and 39 hormone-responsive elements. The analysis identified a total of 153 cis-acting elements belonging to 10 different functional types in the VrPEBP genes. It is noteworthy that there were variations in both the function and distribution of these cis-elements within each gene (Fig. 5). VrTFL1 and VrFT-like1 promoters stood out by containing the highest number of cis-acting elements, with a total of 31 elements each, while VrFT1 promoters contained the fewest, with only 15 cis-acting elements (Fig. 5a). One striking observation is that all VrPEBP genes featured light-responsive elements, emphasizing the significance of light in their regulation (Fig. 5b). However, differences emerged in the distribution and types of stress-responsive and hormone-responsive elements among these genes. For instance, the majority of VrPEBP genes contained anaerobic-responsive elements, drought-responsive elements, and abscisic acid-responsive elements, with only VrFT-like2 lacking anaerobic-responsive elements, VrMFT1 lacking drought-responsive elements, and VrFT1 lacking abscisic acid-responsive elements. Furthermore, among the stress-responsive elements, anaerobic and drought elements were more widely distributed than low-temperature and other stress-related elements. Among the hormone-responsive elements, elements related to MeJA and abscisic acid were more abundant compared to other hormone-related elements. These findings underscore the prevalence of elements associated with stress resistance and hormonal regulation in the VrPEBP genes, suggesting their vital roles in plant responses to stress and hormonal cues.
Analysis of the cis-acting elements in the promoter regions of 7 PEBP genes. (A) The numbers of cis-acting elements in 7 PEBP genes promoters. (B) Distribution of cis-elements in the promoters of mung bean PEBP genes. Gray lines indicate the promoters. Cis-elements differing in function are color-coded accordingly.
Photoperiod response analysis of PEBP genes in mung bean
The mung bean variety LZL132, grown at the experimental field of the Crop Institute, Liaoning Academy of Agricultural Sciences, started flowering 35 days after emergence. Different tissues at the flowering initiation stage were obtained to perform relative expression level detection of VrPEBP using the qPCR method. The results revealed significant tissue-specificity among different PEBP genes in mung bean plants. For example, VrFT1, a member of the FT subfamily, exhibited the highest expression levels in leaves, while members of the TFL subfamily, VrTFL1, VrTFL2, and VrTFL3, showed significantly higher expression levels in the roots compared to other tissues. Members of the MFT family, such as VrMFT1, displayed higher expression levels in pods, whereas VrFT-like1 and VrFT-like2 exhibited relatively lower expression levels compared to other genes (Fig. 6).
Tissue-specific expression analysis of VrPEBP using qPCR. The data are presented as means ± standard errors (SEs) derived from three independent biological replicates. Statistically significant differences among the different tissues are indicated by different lowercase letters (a, b, c or d) based on Duncan’s test (p-value < 0.05).
The study employed artificial climate chambers to cultivate the plants under two treatments: long-day (LD) and short-day (SD) photoperiods. Under the SD treatment, flower buds appeared and bloomed in 32 days, while under the LD conditions, flower buds appeared and bloomed in 36 days. The number of daily flowers produced was recorded throughout the flowering period. The flowering time of each individual flower was also recorded, and the statistical analysis of the distribution of flowering times revealed that flowering significantly occurred earlier under the SD treatment compared to the LD treatment (Fig. 7a).
The statistical analysis of flowering phenotypes in mung beans under long-day (LD) and short-day (SD) treatments, as well as the expression patterns of the PEBP gene in response to photoperiod. (A) Statistical results of flowering time in mung bean plants (days after emerge of flower, DAE) under LD and SD conditions. The significance test for the difference in flowering time between long-day and short-day conditions is conducted using the Wilcoxon rank-sum method. (B) Expression patterns of the PEBP gene in mung bean plants in response to photoperiod under LD and SD conditions.
Furthermore, the expression levels of the PEBP gene in mung bean leaves under different photoperiods were investigated. Samples were collected at various time points on the 32nd day after emergence under the SD treatment, with corresponding samples collected under the LD treatment as controls. The results indicated that VrFT1 and VrMFT1 exhibited significantly higher expression levels than the LD treatment at 8 h and 4 h, respectively, under SD conditions. However, VrFT-like1, VrFT-like2 and VrTFL1 did not show significant changes in expression levels under the different photoperiod treatments. On the other hand, VrTFL2 and VrTFL3 exhibited significantly higher expression levels than the SD treatment at 8 h and 12 h, respectively, under the LD conditions. These findings suggest that VrFT1 and VrMFT1 promote flowering under SD conditions, while VrTFL2 and VrTFL3 inhibit flowering in mung bean plants under SD conditions (Fig. 7b).
Subcellular localization of VrFT1
In silico predictions suggested the cytoplasmic localization of VrFT1. To validate this, we conducted RT-PCR to clone VrFT1's CDS sequence. Subsequently, a fusion vector comprising VrFT1 and green fluorescent protein (GFP), driven by the CaMV 35S promoter, was constructed. To assess subcellular distribution, we transiently infiltrated N. benthamiana leaves with both the VrFT1::GFP fusion vector and a GFP control. Post-infiltration, GFP control fluorescence permeated the entire cell (Fig. 8), while VrFT1::GFP fluorescence was predominantly observed within the nucleus, with some concentration in the vicinity of the cytoplasm near the nucleus (Fig. 8). These findings suggest that VrFT1 is present and operational in both the cytoplasm and the nucleus.
VrFT1 promotes flowering in transgenic Arabidopsis plants
The VrFT1 gene exhibits higher expression levels under short-day treatment and promotes flowering. To investigate the function of VrFT1, we transformed it with the 35S promoter into Columbia-type Arabidopsis and obtained transgenic plants of the T3 generation through subsequent PCR identification. Compared to the wild type, the transgenic Arabidopsis plants showed early flowering (Fig. 9a). The 35S::VrFT1 plants flowered at 33 days (Fig. 9b). The wild-type flowering time was 43 days, while the VrFT1 overexpressing plants flowered at 39 days. However, there was no significant change in the number of rosette leaves. These results indicate that VrFT1 can promote flowering without significantly altering the process of nutrient production. To further identify whether VrFT1 can regulate flowering-related genes in Arabidopsis, AtAP1, AtFUL, AtSOC1, and AtLFY were selected for analysis. The expression level of VrFT1 was significantly higher than that of Col-0 in the overexpressing Arabidopsis plants (Fig. 9c). Among the flowering-related genes, AtAP1, AtFUL, and AtSOC1 were significantly upregulated in the VrFT1 overexpressing plants compared to the wild type.
Flowering-related traits and gene expression in Arabidopsis plants overexpressing the VrFT1 Gene. (A) Differential flowering phenotypes between Arabidopsis plants overexpressing 35S::VrFT1 and wild-type Columbia (Col-0) under long-day conditions. (B) Statistical analysis of flowering-related traits in Col-0 and transgenic Col-0 Arabidopsis. (C) Expression levels of flowering-related genes in Arabidopsis plants overexpressing the VrFT1 gene and their wild-type counterparts. Values are presented as means ± standard deviation (SD). Different letters within the same column indicate significant differences (P < 0.05).
Discussion
Flowering time is a crucial stage where vegetative growth shifts towards reproductive growth, directly impacting crop yield and quality32,33,34Studying the flowering time and its molecular mechanisms in crops is of great importance for understanding crop yield and quality formation35. As a key gene family involved in the regulation of flowering, the function of PEBP genes in mung bean has not been investigated. PEBPgenes of some leguminous crops have been successfully identified, but the specific functions of most of these genes are still unclear36. It is particularly worth noting that as an important member of the leguminous family, soybean has made significant progress in research on its PEBPgene, especially in the field of photoperiod-controlled flowering37. In flowering plants, Flowering locus T (FT) gene is involved in the production of florigen molecule which is essential for induction of flowering, influenced largely by the duration of photoperiod38. Research on the soybean PEBP gene shows that GmFT5b is a key factor in soybean flowering, GmFT2a is an integrator of the soybean photoperiodic flowering pathway, and GmFT1ahas a flowering inhibitor effect39,40,41.
Mung bean is a short-day crop, and exploring their flowering period and response to photoperiod is of significant importance for mung bean production42,43. The influence of different latitudes on photoperiod is mainly reflected in the changes in day and night length. Therefore, this study conducted a genome-wide identification, evolutionary analysis, expression pattern characterization, and functional study of PEBP genes in mung bean, with a focus on their response to photoperiod. Investigating the flowering time and its response mechanisms to photoperiod are of significant importance for the introduction and breeding of mung bean. Furthermore, we conducted functional characterization of VrFT1, elucidating its expression patterns under different photoperiod conditions and its role in promoting flowering. The expression levels identified in this study can serve as biomarkers for detecting the flowering status in different mung bean varieties, thereby providing an effective tool for breeding mung bean varieties adapted to different latitudes.
In this study, seven genes containing the complete PEBP conserved domain were identified in the reference genome of mung bean. However, the distribution of these genes on the chromosomes was not uniform, with only Vr01, Vr03 and Vr08 being identified as PEBP genes on the 11 chromosomes of mung bean (Fig. 1)44. The VrPEBP genes were classified into four different types, including FT, TFL, MFT and FT-like. The TFL and FT subfamilies exhibited a similar gene structure to PEBP genes in other plants, with four exons and three introns, and the two outer exons were longer compared to the two middle exons. This indicates that the VrPEBPgene structure in the FT and TFL subfamilies is relatively conserved and may have similar functions to FT and TFL in other plants5. The sole member of the MFT subfamily, VrMFT1, had nine exons and eight introns, which differed from the characteristic four exons and three introns found in other species. However, the five exons located at the 3’-terminal did not contain the PEBP domain. Further in-depth research is needed to determine whether these exons have functional implications for VrMFT1. Although two genes in the FT-like subgroup were identified to have the complete PEBP domain, their gene structure and motif composition differed significantly from other PEBP genes. Furthermore, their expression levels were low in different tissues and induced by photoperiod, suggesting that they may be pseudogenes generated during the evolutionary process (Figs. 6, 7). In terms of motif composition, except for the FT-like genes, the FT, TFL and MFT subgroups exhibited similar motif compositions, with Motif 1 and Motif 3 containing the conserved motifs DPDxP and GxHR, which are crucial sites in the structure of PEBP genes (Fig. 3b, Figure S1). The DPDxP and GxHR motifs are two highly conserved short sequences found in PEBP proteins across different species. They are part of the conserved domain that forms a pocket-like structure responsible for ligand binding. Specific amino acid residues within the DPDxP and GxHR sequences may influence the function and interactions of PEBP proteins45. Therefore, the PEBP gene sequences in mung bean exhibit a relatively conserved structure, with FT and TFL type genes sharing similar structural characteristics to other crops, while the VrPEBP gene structures in the MFT and FT-like subgroups differ from those in other species.
Gene duplication can increase the diversity and complexity of plant genomes, providing more possibilities for plants to adapt to different environments and stresses, such as the formation of floral organs, induction of disease resistance, and regulation of stress responses46. Mung bean exhibits a rich presence of collinear modules throughout the entire genome, but the PEBPgenes in this study were not found in any of these modules, indicating the absence of segmental or tandem duplications during the evolutionary process of the mung bean genome. This may be related to the fact that the mung bean genome has not undergone whole-genome duplication and is relatively small, with only 441 Mb31. The results of collinearity analysis with other species showed that the VrPEBP genes mainly originated from the retention of orthologous genes during species evolution (Fig. 4b). The fact that mung bean shares more PEBP genes with other legume crops such as peanuts and soybeans than with dicotyledonous plant Arabidopsis and even more than with monocotyledonous plants like maize and rice supports this result.
Tissue-specific expression analysis revealed differential expression of VrPEBP in different tissues. On the other hand, VrMFT1 shows higher expression levels in pods. MFT genes play crucial roles in seed formation, development, and germination processes 47. MFT in wheat is a negative regulator of seed germination and temperature-dependent seed dormancy, and that a single nucleotide polymorphism in the MFTpromoter may be responsible for the natural variation of seed dormancy48. Overexpression of GmMFT affects seed oil and protein content and seed weight in both soybean and Arabidopsis49. MFT genes represses seed germination under far-red light conditions in Arabidopsis thaliana, and function analysis reveals that MFTmodulates the levels and responses of two phytohormones: abscisic acid (ABA) and gibberellin (GA), which have opposite effects on seed germination16. In this study, VrMFT also exhibited high expression in seeds, indicating its significant role in the seed formation process of mung beans. Additionally, cis-element analysis revealed that the VrMFT gene contains regulatory elements associated with hormones such as MeJA, ABA and auxin, suggesting that VrMFT may participate in the regulation of these hormone types, thereby contributing to seed formation and germination processes. However, the three genes of the TFL subfamily exhibited notably higher expression levels in roots compared to other tissues. TFL, as a flowering suppressor protein, plays a role in inhibiting flowering in Arabidopsis. While the expression of TFL-type genes in roots has been detected in many studies, there is limited research on their functions related to roots50,51. Therefore, the involvement of TFL genes in root-related functions in mung beans may suggest that they have novel biological functions.
Light-responsive elements such as G-BOX, Box 4, and GT1 are significantly enriched in the promoter region of VrPEBP compared to other types (Table S5), suggesting that the VrPEBP gene primarily participates in the light-responsive molecular mechanisms in mung bean. Mung bean, a short-day plant, exhibit a significant delay in flowering under long-day conditions (Fig. 7a). The FT and TFL gene types show opposite patterns in response to photoperiod. VrFT1 gene expression is upregulated under short-day conditions but lower under long-day conditions, while the VrTFL2 and VrTFL3 genes of the TFL subfamily are expressed at higher levels under long-day conditions. This indicates that VrFT1 promotes flowering, while VrTFL inhibits flowering under short-day conditions. To further validate its flowering promotion function, VrFT1 was overexpressed in transgenic Arabidopsis plants, and the results showed that it could promote early flowering in Arabidopsisunder LD conditions. The highest Hd3a of rice expression was observed in leaves when exposed to short-day (SD) conditions25,52. Likewise, in the long-day (LD) plant Arabidopsis, the leaves exhibited the highest expression levels under LD conditions9. This pattern was also observed in FT genes, indicating a close relationship with seasonal flowering in species such as soybean (Glycine max), poplar (Populus deltoides), and sugarcane (Saccharum spp.) 53–55. Our results revealed a distinct diurnal rhythm in the expression of VrFT1 in mung bean under both SD and LD conditions. The FT-FD module plays a crucial role in the photoperiodic flowering pathway 56. This conceptual framework illustrates the movement of the FT protein within the plant’s vascular system towards the apex57. Upon reaching the apex, the FT protein interacts with the FD transcription factor, a basic Leu zipper protein (TF), to initiate the activation of floral meristem identity genes, including AP1 and FUL58,59,60. Consequently, the FT-FD complex facilitates the expression of flowering-related genes, such as FRUITFULL (FUL), LEAFY (LFY), APETALA1 (AP1) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1)58,59. In this study, the overexpression of VrFT1 in Arabidopsis plants resulted in significantly increased levels of AtAP1, AtFUL, and AtSOC1 compared to the wild-type variety. However, it is worth noting that the LFY assumes a pioneering role in determining floral fate in Arabidopsis. In response to both internal and external stimuli, plants produce specific floral inducers known as florigens. Interestingly, in this study, the expression level of AtLFY was found to be lower in the overexpressed plants compared to the wild-type, possibly due to functional differences arising from variations in the sequence of VrFT1 and AtFT genes in Arabidopsis.
Conclusion
A large amount of literature has demonstrated that the PEBP gene has an important role in controlling flowering in legumes. We therefore provide an in-depth analysis of the regulation of environmental signals affecting flowering. As an important environmental factor, photoperiod has a significant impact on plant growth and development, especially flowering time. Different crops have different sensitivities to photoperiod, which determines their adaptability and yield performance in different regions. Therefore, analyzing and identifying the function of the VrPEBP gene and understanding the photoperiod characteristics of different latitudes will help breeders to carry out targeted modifications of the gene through gene editing, transgenic and other technical means to cultivate plants that adapt to the photoperiod conditions in specific regions. The PEBPgene family is a key marker gene controlling flowering in various crops, with a directional effect on crop flowering. Although reports have identified genes related to flowering time in mung beans through QTL and GWAS analyses, no PEBP gene family members were found within the targeted intervals of these studies62.
In our study, a comprehensive in silico analysis was conducted to investigate the VrPEBP gene family in mung bean, including evolutionary analysis, gene structure examination and identification of regulatory elements. A total of 7 VrPEBP genes were identified and classified into four groups. Differential expression patterns of VrPEBP genes in different tissues and under different photoperiodic treatments were analyzed. VrFT1 was chosen for functional validation in transgenic Arabidopsis plants. It was found to VrFT1 gene exhibits higher expression levels under short-day treatment and promotes flowering. These findings provide valuable insights into the functional characterization of VrPEBP genes and will enable the development of functional markers based on the gene sequences of these confirmed functional genes, which can be used for molecular marker-assisted breeding.
Materials and methods
Plant materials and treatment
The plant material used in this study, LZL132, with an early flowering genotype in mung beans, was provided by the Crop Research Institute of the Liaoning Academy of Agricultural Sciences. Its flowering period occurs approximately 35 days after emergence and is a widely cultivated mung bean material in the northeastern region of China. To identify the tissue-specific expression of the VrPEBP gene, root, stem, leaf, flower, and pod tissues of mung bean were collected after flowering in the field environment and stored at −80 °C. To identify the response of the VrPEBP gene under different photoperiod treatments, mung bean seeds were sown in pots with a 25-cm diameter containing a mixture of nutrient soil and vermiculite (1:1 ratio). The pots were placed in a growth chamber at a temperature of 26 ℃. Two photoperiods were set: long-day conditions LD (14 h of light and 10 h of darkness) and short-day conditions SD (10 h of light and 14 h of darkness). The time of flower bud differentiation and flowering in mung bean plants was recorded. In LZL132, flower buds appeared after 33 days, and within 24 h of bud appearance, leaf samples were collected at six different time points: 0 h, 4 h, 8 h, 12 h, 16 h, and 20 h. At the same time points, leaf samples were also collected from mung bean plants grown under long-day conditions at the same location for comparison. All collected tissues were stored at −80 ℃ for future use.
Identification of PEBP family members in mung bean
The mung bean genome data were obtained from the website the Legume Information System https://www.legumeinfo.org/31. Initially, the consensus seed file (PF01161) for the hidden Markov model (HMM) was downloaded from the Pfam website. Subsequently, the HMM profile was employed as a query to detect PBP-containing domains in the mung bean genome, using a threshold e-value of < e–5. A similar approach was used to identify PEBP genes in Phaseolus vulgaris, Cicer arietinum, Zea mays, Sorghum bicolor, Vitis vinifera, and Medicago truncatula. Information about PEBP family membership in soybean, cultivated peanut, rice, and Arabidopsis was obtained from published articles. Genome sequences and CDS files for various plant species were obtained from the Phytozome database. The potential PEBP protein sequences underwent analysis on SMART (https://smart.embl.de/) and NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/) to identify PEBP domains and remove non-representative transcripts. Redundant sequences or those with incomplete PEBP domains were excluded from subsequent analyses. The protein sequences of identified PEBP gene family members were subjected to the Expasy website (https://web.expasy.org/) for calculating theoretical pI and molecular weight. To determine the subcellular location of VrPEBPs, the PSORT website (https://wolfpsort.hgc.jp) was utilized.
Gene structure and motif prediction of VrPEBP genes
The annotation files for PEBP gene family members were sourced from the Legume Information System (https://www.legumeinfo.org/). To depict the functional structural domains of PEBP family proteins, NCBI’s CDD (Conserved Domain Database, https://www.ncbi.nlm.nih.gov/cdd/) was utilized. For the identification of conserved motifs within mung bean’s PEBP protein sequences, online MEME software (http://meme-suite.org/tools/meme) was employed with a motif count of 7, leaving the other parameters as default conditions. Lastly, TBtools was used to facilitate the visualization of gene evolutionary trees, gene structures, as well as domains and conserved motifs.
Chromosomal distribution analysis of VrPEBP genes
According to the information within the the Legume Information System (https://www.legumeinfo.org/), the specific positions of the PEBPgene family members on the chromosomes of mung bean were analysed. Then, TBtools software was used to map these genes to individual chromosomes63.
Evolutionary and homology analysis of VrPEBP gene family
To explore the divergence and evolutionary relationships of mung bean’s PEBP gene family with those of other plant species, we constructed a phylogenetic tree. This tree encompassed protein sequences from VrPEBP genes and PEBP family genes from species such as Arabidopsis thaliana, Phaseolus vulgaris, Glycine Max, Cicer arietinum, cultivated peanuts, Oryza Sativa, Zea mays, Sorghum bicolor, Vitis vinifera, and Medicago truncatula. We aligned the PEBP protein sequences of these eleven species, including mung bean, using the ClusterW method with default parameters in MEGA11 software64,65. For tree construction, we applied the Neighbor-Joining (NJ) method, employing a bootstrap method with 1000 replicates and the Poisson model. Subsequently, the EvolView website (www.evolgenius.infi/evolview/#/terrview) was utilized to enhance the tree’s visual presentation66. Genomic data in FASTA format and GFF3 gene annotation files for Arabidopsis thaliana, Phaseolus vulgaris, Glycine Max, Cicer arietinum, cultivated peanuts, Oryza Sativa, Zea mays, Sorghum bicolor, Vitis vinifera, and Medicago truncatula were downloaded from the Phytozome Database (https://phytozome-next.jgi.doe.gov/). Using the genome sequence and gene annotation files, we employed the one-step MCScanX plug-in in TBtools to generate collinearity files between pairs of species, including collinearity and collinear gene pairs. The Ka and Ks values of the obtained orthologous gene pairs were used to calculate the Ka/Ks ratio for all homologous gene pairs67.
Analysis of cis-acting elements in the VrPEBP promoter
The 2000 bp sequence upstream of the VrPEBP genes was extracted using TBtools to analyses cis-acting elements, sequences were organized in one text file, and then PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to identify cis-acting elements. TBtools was then used to visualize the results.
RNA extraction and quantitative real-time PCR
Plant tissues were processed for total RNA extraction with the Plant Total RNA Extraction Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s guidelines. Subsequently, cDNA was synthesized using the PrimeScript™ RT Kit (TaKaRa, Japan). The reference gene selected was Actin, and the corresponding primers were designed via Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). For gene expression analysis, the SYBR Premix Ex TaqII kit (TliRNaseH Plus) from TaKaRa, Japan, was employed, and fluorescence quantitative reactions were detected using Applied Biosystems’ ABI7500 instrument. Relative expression analysis was computed using the 2−ΔΔCT method.
Subcelluar localization analysis.
The complete cDNA of VrFT1 was amplified from mung bean leaves using specific primers. The coding sequence data for VrFT1 were obtained from the Legume Information System for reference. The resulting PCR products were ligated into the pBWA (V) HS-GLosgfp vector, generating the pBWA (V) HS-VrFT1-GFP vector, which contained the reporter gene for green fluorescent protein (GFP). After confirming the sequences through sequencing, the positive clones were transformed into Agrobacterium tumefaciens (EHA105) using electrotransformation. Carefully selected tobacco seedlings (30 days old) were injected in the lower epidermis of their leaves. Subsequently, these seedlings were cultured under low light conditions for a period of 2 days. Observation and imaging were performed using a laser confocal microscope (Nikon C2-ER, Tokyo, Japan), with the corresponding empty vector used as a control.
The VrFT1 function analysis in transgenic Arabidopsis under different photoperiod
To initiate the amplification of VrFT1 cDNA in LZL132, we designed gene-specific primers using Primer Premier 6.0. The PCR amplification of the coding sequence was carried out using the TransTaq DNA Polymerase High Fidelity Amplification Kit. The PCR program involved an initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58–60 °C for 30 s, and extension at 72 °C for 1 min. Finally, a 10-min extension at 72 °C concluded the process. The ligated DNA was then introduced into Escherichia coli Top10 competent cells, where positive clones were selected and their identity confirmed through sequencing. Following this, the recombinant plasmid pBWA (V) BS-VrFT1 was introduced into Agrobacterium tumefaciens EHA105 and subsequently used to transform wild-type Arabidopsis (Col-0) through the floral dip. After screening with antibiotics and PCR verification of the transgenic seedlings, homozygous transgenic lines were successfully generated in the T2 generation. Subsequently, homozygous T3 progeny were scrutinized and selected for further experimental procedures. T3 seeds, which had previously undergone a 4 °C vernalization treatment on MS medium, were then incubated in a growth chamber at 22 °C for 14 days. Subsequently, Arabidopsis seedlings were transplanted into plastic pots containing a nutrient soil and vermiculite mixture (3:1 ratio) for further cultivation. The plants were subjected to long-day conditions with a photoperiod of 16 h of daylight and 8 h of darkness, a photosynthetic photon flux density of 400 μmol m − 2 s − 1, and a relative humidity of 70%. After 14 days of transplantation, the expression levels of VrFT1 and its associated flowering-related genes were investigated. Columbia wild-type Arabidopsis plants (Col-0) were used as controls and subjected to the same growth conditions.
Statistical analysis
In the case of multiple comparisons between different groups, Duncan’s test, the Wilcoxon rank-sum method and means ± standard deviation (SD)was used. All data were analyzed using R language (version 4.3.0), and all statistical analyses in this study were conducted using the respective R packages. All the values were calculated as the means ± standard deviation (SD). Asterisks indicate significant differences. *P < 0.05. **P < 0.01. Error bars represent the standard deviation from triplicate values.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors. The VrFT sequences reported in this paper have been submitted to NCBI (https://www.ncbi.nlm.nih.gov/) GenBank database under the accession numbers PP693231. No endangered plant species were used and this experiment complied with relevant institutional, national and international guidelines and legislation.
References
Fornara, F., de Montaigu, A. & Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 141, 550-550.e552. https://doi.org/10.1016/j.cell.2010.04.024 (2010).
Kinoshita, A. & Richter, R. J. J. o. e. b. Genetic and molecular basis of floral induction in Arabidopsis thaliana. 71, 2490–2504 (2020).
Bao, S., Hua, C., Shen, L. & Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. Journal of Integrative Plant Biology 62, 118–131. https://doi.org/10.1111/jipb.12892 (2020).
Huang, X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics 44, 32–39. https://doi.org/10.1038/ng.1018 (2011).
Karlgren, A. et al. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiology 156, 1967–1977. https://doi.org/10.1104/pp.111.176206 (2011).
Freytes, S. N., Canelo, M. & Cerdán, P. D. Regulation of Flowering Time: When and Where? Current Opinion in Plant Biology 63, https://doi.org/10.1016/j.pbi.2021.102049 (2021).
Liu, Y. Y., Yang, K. Z., Wei, X. X. & Wang, X. Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytologist 212, 730–744. https://doi.org/10.1111/nph.14066 (2016).
Putterill, J. & Varkonyi-Gasic, E. FT and florigen long-distance flowering control in plants. Current Opinion in Plant Biology 33, 77–82. https://doi.org/10.1016/j.pbi.2016.06.008 (2016).
Luo, X., Chen, T., Zeng, X., He, D. & He, Y. Feedback Regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5′ FLC Promoter Region in Arabidopsis. Molecular Plant 12, 285–288. https://doi.org/10.1016/j.molp.2019.01.013 (2019).
Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M. & Araki, T. TWIN SISTER OF FT (TSF) Acts as a Floral Pathway Integrator Redundantly with FT. Plant and Cell Physiology 46, 1175–1189. https://doi.org/10.1093/pcp/pci151 (2005).
Huang, N. C., Jane, W. N., Chen, J. & Yu, T. S. Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. The Plant Journal 72, 175–184. https://doi.org/10.1111/j.1365-313X.2012.05076.x (2012).
Yoo, S. J. et al. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. The Plant Journal 63, 241–253. https://doi.org/10.1111/j.1365-313X.2010.04234.x (2010).
Wang, Z. et al. The Divergence of Flowering Time Modulated by FT/TFL1 Is Independent to Their Interaction and Binding Activities. Frontiers in Plant Science 8, https://doi.org/10.3389/fpls.2017.00697 (2017).
Jin, S., Nasim, Z., Susila, H. & Ahn, J. H. Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Seminars in Cell & Developmental Biology 109, 20–30. https://doi.org/10.1016/j.semcdb.2020.05.007 (2021).
Ryu, J. Y. et al. The Arabidopsis Floral Repressor BFT Delays Flowering by Competing with FT for FD Binding under High Salinity. Molecular Plant 7, 377–387. https://doi.org/10.1093/mp/sst114 (2014).
Vaistij, F. E. et al. MOTHER-OF-FT-AND-TFL1
represses seed germination under far-red light by modulating phytohormone responses in
Arabidopsis thaliana. Proceedings of the National Academy of Sciences 115, 8442–8447, https://doi.org/10.1073/pnas.1806460115 (2018).
Angeles-Shim, R. B. et al. Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. Plos One 14, https://doi.org/10.1371/journal.pone.0215771 (2019).
Sun, Y. et al. Genome-Wide Identification of PEBP Gene Family in Solanum lycopersicum. International Journal of Molecular Sciences 24, https://doi.org/10.3390/ijms24119185 (2023).
Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T. & Handa, H. Molecular and Functional Characterization of PEBP Genes in Barley Reveal the Diversification of Their Roles in Flowering
Plant Physiology 149, 1341–1353, https://doi.org/10.1104/pp.108.132134 (2009).
Carmona, M. J., Calonje, M. & Martínez-Zapater, J. M. The FT/TFL1 gene family in grapevine. Plant Molecular Biology 63, 637–650. https://doi.org/10.1007/s11103-006-9113-z (2006).
Zhao, C. et al. Genomic Survey of PEBP Gene Family in Rice: Identification, Phylogenetic Analysis, and Expression Profiles in Organs and under Abiotic Stresses. Plants 11, https://doi.org/10.3390/plants11121576 (2022).
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V. & Simmons, C. R. A Genomic and Expression Compendium of the Expanded PEBP Gene Family from Maize. Plant Physiology 146, 250–264. https://doi.org/10.1104/pp.107.109538 (2008).
Zhong, C. et al. Comparative Genomic and Expression Analysis Insight into Evolutionary Characteristics of PEBP Genes in Cultivated Peanuts and Their Roles in Floral Induction. International Journal of Molecular Sciences 23, https://doi.org/10.3390/ijms232012429 (2022).
Song, C., Li, G., Dai, J. & Deng, H. Genome-Wide Analysis of PEBP Genes in Dendrobium huoshanense: Unveiling the Antagonistic Functions of FT/TFL1 in Flowering Time. Frontiers in Genetics 12, https://doi.org/10.3389/fgene.2021.687689 (2021).
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S. & Shimamoto, K. Hd3a Protein Is a Mobile Flowering Signal in Rice. Science 316, 1033–1036. https://doi.org/10.1126/science.1141753 (2007).
Liu, W. et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytologist 217, 1335–1345. https://doi.org/10.1111/nph.14884 (2017).
Meng, X., Muszynski, M. G. & Danilevskaya, O. N. TheFT-LikeZCN8Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize The Plant Cell 23, 942–960, https://doi.org/10.1105/tpc.110.081406 (2011).
Sengupta, K. in Forage Crops of the World, Volume II: Minor Forage Crops 159–174 (Apple Academic Press, 2018).
Nair, R., & Schreinemachers, P. Global status and economic importance of mungbean. The mungbean genome, 1–8 (2020).
Dahiya, P. K. et al. Mung Bean: Technological and Nutritional Potential. Critical Reviews in Food Science and Nutrition 55, 670–688. https://doi.org/10.1080/10408398.2012.671202 (2014).
Liu, C. et al. High-quality genome assembly and pan-genome studies facilitate genetic discovery in mung bean and its improvement. Plant Communications 3, https://doi.org/10.1016/j.xplc.2022.100352 (2022).
Han, T. et al. Postflowering photoperiod regulates vegetative growth and reproductive development of soybean. Environmental and Experimental Botany 55, 120–129. https://doi.org/10.1016/j.envexpbot.2004.10.006 (2006).
Khan, M. R. G., Ai, X. Y. & Zhang, J. Z. Genetic regulation of flowering time in annual and perennial plants. WIREs RNA 5, 347–359. https://doi.org/10.1002/wrna.1215 (2013).
González, A. M. et al. Major Contribution of Flowering Time and Vegetative Growth to Plant Production in Common Bean As Deduced from a Comparative Genetic Mapping. Frontiers in Plant Science 7, https://doi.org/10.3389/fpls.2016.01940 (2016).
Jung, C. & Müller, A. E. Flowering time control and applications in plant breeding. Trends in Plant Science 14, 563–573. https://doi.org/10.1016/j.tplants.2009.07.005 (2009).
Zhong, Chao. et al. Comparative genomic and expression analysis insight into evolutionary characteristics of PEBP genes in cultivated peanuts and their roles in floral induction. International Journal of Molecular Sciences 23.20.12429 (2022).
Cai, Zhandong. et al. MOTHER‐OF‐FT‐AND‐TFL1 regulates the seed oil and protein content in soybean. New Phytologist 239, 905–919 (2023).
Tribhuvan, Kishor U. et al. Identification and characterization of PEBP family genes reveal CcFT8 a probable candidate for photoperiod insensitivity in C. cajan. 3 Biotech 10, 1–12 (2020).
Su, Qiang, et al. The FLOWERING LOCUS T 5b positively regulates photoperiodic flowering and improves the geographical adaptation of soybean. Plant, Cell & Environment 47, 246–258 (2024).
Cai, Yupeng. et al. CRISPR/Cas9‐mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant biotechnology journal 16, 176–185 (2018).
Liu, Wei, et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytologist 217, 1335–1345 (2018).
Imrie, B. & Lawn, R. J. E. a. Time to flowering of mung bean (Vigna radiata) genotypes and their hybrids in response to photoperiod and temperature. 26, 307–318 (1990).
Wang, N. et al., https://doi.org/10.21203/rs.3.rs-24572/v1 (2020).
Liu, C., Liu, Q. & Mou, Z. A direct link between BR and SA signaling: Negative regulation of TGA4 by BIN2. Mol Plant 15, 1254–1256. https://doi.org/10.1016/j.molp.2022.06.006 (2022).
Yang, Z. et al. Identification and Characterization of the PEBP Family Genes in Moso Bamboo (Phyllostachys heterocycla). Scientific Reports 9, https://doi.org/10.1038/s41598-019-51278-7 (2019).
Panchy, N., Lehti-Shiu, M. & Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiology 171, 2294–2316. https://doi.org/10.1104/pp.16.00523 (2016).
Hedman, H., Källman, T. & Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Molecular Biology 70, 359–369. https://doi.org/10.1007/s11103-009-9478-x (2009).
Nakamura, S. et al. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. 23, 3215–3229 (2011).
Cai, Z. et al. MOTHER-OF-FT-AND-TFL1 regulates the seed oil and protein content in soybean. New Phytologist 239, 905–919. https://doi.org/10.1111/nph.18792 (2023).
Jin, H. et al. Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biology 19, https://doi.org/10.1186/s12870-019-2113-3 (2019).
Adeyemo, O. S., Hyde, P. T. & Setter, T. L. Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reproduction 32, 181–191. https://doi.org/10.1007/s00497-018-00354-5 (2018).
Tamaki, S. et al. FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proceedings of the National Academy of Sciences 112, https://doi.org/10.1073/pnas.1417623112 (2015).
Lin, X., Liu, B., Weller, J. L., Abe, J. & Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. Journal of Integrative Plant Biology 63, 981–994. https://doi.org/10.1111/jipb.13021 (2021).
Zhao, T. et al.Identification of key flowering-related genes and their seasonal expression in Populus tomentosa reproductive buds suggests dual roles in floral development and dormancy. 161, 113175 (2021).
Venail, J. et al. Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS T orthologue in sugarcane. Journal of Experimental Botany 73, 2035–2049. https://doi.org/10.1093/jxb/erab539 (2022).
Li, D. et al. Arabidopsis Class II TCP Transcription Factors Integrate with the FT–FD Module to Control Flowering. Plant Physiology 181, 97–111. https://doi.org/10.1104/pp.19.00252 (2019).
Turck, F., Fornara, F. & Coupland, G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annual Review of Plant Biology 59, 573–594. https://doi.org/10.1146/annurev.arplant.59.032607.092755 (2008).
Abe, M. et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. 309, 1052–1056 (2005).
Wigge, P. A. et al. Integration of Spatial and Temporal Information During Floral Induction in
Arabidopsis. Science309, 1056–1059, https://doi.org/10.1126/science.1114358 (2005).
Jung, J.-H., Lee, H.-J., Ryu, Jae Y. & Park, C.-M. SPL3/4/5 Integrate Developmental Aging and Photoperiodic Signals into the FT-FD Module in Arabidopsis Flowering. Molecular Plant 9, 1647–1659, https://doi.org/10.1016/j.molp.2016.10.014 (2016).
Seo, H. et al. Identification of novel loci for flowering time in mungbean [Vigna radiata (L.) R. Wilczek] using genome-wide association study. Genes & Genomics, 45, 911–919 (2023).
Chiteri, K. O. et al. Combining GWAS and comparative genomics to fine map candidate genes for days to flowering in mung bean. BMC genomics 25, 270 (2024).
Chen, C. et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948, https://doi.org/10.1093/bioinformatics/btm404 (2007).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38, 3022–3027. https://doi.org/10.1093/molbev/msab120 (2021).
He, Z. et al. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. 44, W236-W241 (2016).
Wang, D., Zhang, Y., Zhang, Z., Zhu, J. & Yu, J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80, https://doi.org/10.1016/S1672-0229(10)60008-3 (2010).
Funding
This research was funded by the Project of Shenyang Youth Science and Technology Innovation Talent Supporting Plan (RC220453), Youth Top Talent Project of Liaoning Revitalization Talent Plan (3203201), China Agriculture Research System of MOF and MARA-Food Legumes (CARS-08-Z07), Disciplinary construction project of Liaoning Academy of Agricultural Sciences (2022DD030905) and National Natural Science Foundation of China (31972962).
Author information
Authors and Affiliations
Contributions
XR, YL, CZ and WG contributed to conception and design of the study. YL and YH conducted the bioinformatics analysis. YZ and MF analyzed the data. JC and TL performed the statistical analysis. CZ and YL wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xue, R., Liu, Y., Feng, M. et al. Genome-wide characterization of PEBP genes in Mung bean (Vigna radiata L.) with functional analysis of VrFT1 in relation to photoperiod. Sci Rep 14, 26413 (2024). https://doi.org/10.1038/s41598-024-73936-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73936-1