Abstract
Hepatitis B is one of the public health priorities worldwide, especially in the Southwest China. Our study aimed to investigate the relationship between genotypes and drug resistance mutations among HBV patients in Southwest China, with the objective of providing guidance for clinical antiviral treatment. A total of 4266 chronic hepatitis B (CHB) patients treated in the Qianjiang Hospital of Chongqing University were included in our study from 2014 to 2020. Both genotypes and drug-resistant mutations of CHB patients were determined by polymerase chain reaction (PCR). Genotype B and genotype C were the main HBV genotypes in our study. We found 54 mutation patterns, including 9 single-site mutations and 45 multiple-site mutations, accounting for 57.64% and 42.36%, respectively. rtM204I/V/S (485/1936) was the most common single-site mutation type, and rtL180M + rtM204I/V (482/1936) was the most common multiple-site mutation type. 1372 CHB patients were resistant to LAM + LDT, and 342 CHB patients were resistant to ADV. There was only 1 CHB patient who exhibited resistance to LAM + LDT + ADV + ETV, with a specific mutation pattern of rtA181T + rtT184L + rtM204V. Our study demonstrated trends in genetic mutations and drug resistance in CHB patients to enable timely adjustment of antiviral treatment strategies.
Similar content being viewed by others
Introduction
Hepatitis B virus (HBV) infection remains a major public health problem, causing an estimated 686,000 deaths per year worldwide. In China, it is one of the leading causes of mortality, with over 300,000 deaths per year primarily attributed to cirrhosis and hepatocellular carcinoma1,2,3. HBV, a DNA virus belonging to the Hepadnaviridae family, is characterized by a partially double-stranded DNA genome of approximately 3.2 kb in length. This viral genome encodes several proteins, including HBsAg, HBcAg, HBeAg, viral polymerase, and HBx protein4. At present, HBV is known to have at least 9 genotypes (A to I) and 1 undetermined genotype (J)5. Certain genotypes can be divided into several subtypes. HBV genotypes were demonstrated to correlate with various liver diseases and the severity of oxidative damage in patients with HBV-induced liver disease6,7,8. The prevalence of HBV genotypes shows distinct clinical characteristics, indicating that it is a determinant of the outcome of acute or chronic HBV infection9,10,11,12. Therefore, the detection of HBV genotypes is valuable for predicting the efficacy of interferon treatment and evaluating the prognosis of liver disease13,14,15,16.
Genotype B and genotype C are the main HBV genotypes in China. Specifically, genotype B is predominant in Southern China, while genotype C prevails in Northern provinces17,18. Compared to genotype B, genotype C showed high HBV replication capacity, with high levels of HBV DNA in the serum19,20. In addition, the tendency of chronicity was higher and more frequently developed into liver cirrhosis (LC) and hepatocellular carcinoma (HCC) in patients with genotype C than genotype21,22.
HBV genotype and disease progression are related to interferon α (IFN-α) therapy response23,24,25. HBV is a highly mutable virus. The absence of proofreading capability in RNA polymerase and reverse transcriptase (RT) can result in nucleotide variation in the process of reverse transcription and replication. These variations can occur naturally during chronic persistent infection or be induced by antiviral drug treatment. Studies have found a correlation between HBV mutant variants and reduced sensitivity to antiviral drug26,27,28. In antiviral therapy, genotypic resistance refers to the gene mutation of the HBV to acquire resistance to the drugs administered, while reduced sensitivity to antiviral drugs without any genetic mutation is known as phenotypic resistance. The drug resistance mutation of one antiviral drug also appears resistant to two or more antiviral drugs, called cross-resistance. Resistance to at least two different classes of nucleoside(acid) analogs is called multidrug resistance.
In recent years, the prevalence of drug-resistant mutations of HBV has been reported in different regions of China29,30,31,32,33; however, there is a scarcity of large-scale studies on drug-resistant mutations in Southwest China. This study analyzed the HBV genotypes and drug resistance mutation sites of patients with chronic HBV infection in Southwest China, aiming to determine the occurrence of drug resistance and promptly adjust treatment plans according to the detection results.
Results
Clinical characteristics of chronic hepatitis B patients
Chronic hepatitis B (CHB) was most prevalent in patients aged 21–60 years. There were 3319 (77.80%) males and 947 (22.20%) females with an average of 36.4 ± 11.7 years in this study as shown in Table 1. Among 4266 CHB patients, 1936 (45.38%) had mutations in the HBV RT region. We further analyzed the correlation between existing mutations and clinical features, including gender, age, and HBV genotype, among these CHB patients. As shown in Table 1, genotype B and genotype C were the main HBV genotypes in our study, and there were statistically significant differences between the incidence of mutations and age (p < 0.001) and HBV genotype (p = 0.004).
We analyzed the relationship between genotypes and age, gender, and mutation type among 4266 CHB patients. As shown in Table 2 and 2501(58.63%) were infected with HBV genotype B, and 1765(41.37%) with HBV genotype C. Significant differences were found in different genders (P = 0.02), age distribution (P < 0.001), and mutation types (P < 0.001) among CHB patients with these two genotypes. At the same time, we also found that genotype B predominated across groups of different ages, genders, and mutation types.
The constituent ratio of HBV genotypes and genotypic mutation among CHB patients in different years
We conducted a statistical analysis of HBV genotypes and genetypic mutation among CHB patients in Southwest China from 2014 to 2020. From 2015 to 2016, CHB patients with genotype B increased rapidly and remained higher than CHB patients with genotype C (Fig. 1a). In 2018, the prevalence of CHB patients with genotype B reached its peak, accounting for 62.65% (406/648). The highest proportion of CHB patients with genotype C was observed in 2015, accounting for 58.55% (178/304).
The constituent ratio of HBV genotypes and genotypic mutations among CHB patients over the past seven years. (a) Dynamic changes in the constituent ratio of different genotypes among CHB patients over the past seven years; Percentage = cases of CHB patients with genotype B(C) per year/cases of the total number of CHB patients per year. (b) Dynamic changes in the constituent ratio of genotypes among CHB patients with genotypic mutations over the past seven years. Percentage = cases of genotype B(C) patients with genotypic mutations per year/cases of the total number of CHB patients with genotypic mutations per year.
The genotype mutations were detected in 45.38% (1936/4266) of total CHB patients, with genotype B accounting for 25.53% (1089/4266) and genotype C for 19.85% (847/4266). The mutation rate of CHB patients with genotype B and C were 43.54% (1089/2501) and 47.99% (847/1765), respectively. As shown in Fig. 1b, from 2014 to 2020, the tendency in the proportion of genotypic mutations among CHB patients with genotype B and C were consistent with that of HBV genotypes distribution. In 2019, CHB patients with genotype B exhibited the highest proportion of genotypic mutations among all patients with genotypic mutations, accounting for 65.82%. In 2015, CHB patients with genotype C had the highest proportion of genotypic mutations among all patients with genotypic mutations, accounting for 72.06%.
Distribution characteristics of mutation sites of CHB patients with different genotypes
Genotypic mutations were detected in 1936 cases among the 4266 CHB patients, accounting for 45.38% (see Table 2). Next, the mutation site distribution of different HBV genotypic strains in these 1936 CHB patients was further investigated to understand the molecular mechanisms of drug resistance.
Distribution of mutation sites of CHB patients with different HBV genotypes
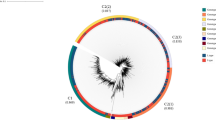
In this study, nucleos(t)ide analogues (NAs) associated mutations were detected at rtV173L, rtL180M, rtA181S/V/T, rtT184I/L, rtS202G, rtM204I/V/S, rtV207I/L/M, rtS213T, rtQ215S, rtN236T/V, and rtM250L/V. Nevertheless, one mutation site without the relationship of NAs resistance was also detected, i.e. rtI187V/L. The mutation sites of the RT region from 1936 CHB patients were described in Table 3. Among the 1936 CHB patients, a total of 2901 mutation sites were detected. The highest frequency of mutations was observed at rtM204I/V/S (1479, 50.67%), followed by rtL180M (629, 21.68%) (Fig. 2a). Among all HBV genotypic mutation sites, genotype B and genotype C accounted for 53.81% and 46.19%, respectively. Among CHB patients with genotypes B and C, rtM204I/V/S was the most common mutation, followed by rtL180M, rtA181S/V/T, rtN236T/V, and rtV207I/L/M (Fig. 2b,c).
The proportion of different mutation sites among CHB patients. (a) The proportion of different mutation sites among the overall HBV mutations; (b), The proportion of different mutation sites among HBV genotype B; (c) The proportion of different mutation sites among HBV genotype C. The pie charts represented the constituent ratio of different mutation sites, with different color representing different mutation sites; Percentage = number of single mutation site in genotype B(C)/the total number of mutation sites in genotype B(C); (d) Dynamic changes in the constituent ratio of primary mutation sites over the past seven years. Percentage = number of single mutation site per year/the total number of mutation sites per year.
Dynamic changes in the constituent ratio of primary mutant sites over the past seven years
The proportion of mutation sites composition also varied across the years from 2014 to 2020 (Fig. 2d). The proportion of rtM204 mutation remained relatively stable from 2014 to 2020, reaching its peak in 2018, when rtM204 mutations accounted for 55.11% of all mutation sites. The proportion of rtL180 mutations remained relatively stable from 2014 to 2020, peaking in 2016, when rtL180 mutations accounted for 23.91% of all mutation sites.
Distribution of drug resistance mutations among CHB patients with different genotypes
In certain cases, mutations were present in combinatorial patterns, resulting in a total of 2901 mutations among 1936 CHB patients. A total of 54 mutation patterns were found in this study, including 9 single-site mutations and 45 multiple-site mutations. CHB patients with single-site mutation accounted for 57.64% (1116/1936), and CHB patients with multiple-site mutations accounted for 42.36% (820/1936) shown as Table 4. The single-site mutations were primarily rtM204I/V/S (752/1936), and secondly rtA181S/V/T (157/1936). Among multiple-site mutations, genotypic mutation at rtL180 + rtM204 (482/1936) was mostly detected, followed by rtA181 + rtN236 (76/1936) and rtM204 + rtV207 (50/1936). Therefore, we classified the mutation patterns according to the expected phenotypes associated with drug resistance (Fig. 3). Single drug resistance was detected in 445 CHB patients (10.43%, 445/4266). Adefovir (ADV) resistance was the most frequently detected single drug resistance (8.02%, 342/4266), followed by lamivudine (LAM) resistance (2.30%, 98/4266) and entecavir (ETV) resistance (0.12%, 5/4266) (Fig. 3a). Multidrug resistance was detected in 1487 CHB patients (34.86%, 1487/4266). The most frequently detected multidrug resistance was lamivudine + telbivudine (LAM + LDT) resistance (32.16%, 1372/4266), followed by LAM + LDT + ETV resistance (1.57%, 67/4266), LAM + LDT + ADV (0.70%, 30/4266), LAM + ADV (0.40%, 17/4266), and LAM + LDT + ADV + ETV resistance (0.02%, 1/4266); no cases of tenofovir resistant strains have been detected (Fig. 3b). In this study, one CHB patient was resistant to LAM + LDT + ADV + ETV, and the corresponding mutation pattern was rtA181T + rtT184L + rtM204V. It was worth noting that there were 4 CHB patients with genotypic mutation unrelated to NAs resistance, and their mutation pattern was rtI187V/L. The RT gene sequence of these four patients were shown in the supplementary Table S1 (Supplementary Materials). These four CHB patients were infected with genotype B. There were 1089 CHB patients of drug resistance infected with genotype B, and the drug resistance rate was 43.54% (1089/2501). Among these 1089 CHB patients of genotype B, the most common single drug resistance was ADV, and the most common multidrug resistance was LAM + LDT. There were 847 CHB patients of drug resistance infected with genotype C, and the drug resistance rate was 47.99% (847/1765). Among these 847 CHB patients of genotype C, the most common single drug resistance was ADV, and the most common multidrug resistance was LAM + LDT.
Distribution of drug resistance patterns among CHB patients with different genotypes in Southwestern China, 2014–2020. (a) Mutation patterns of single drug resistance among CHB patients with different genotypes. (b) Mutation patterns of multidrug resistance among CHB patients with different genotypes.
Dynamic changes of the drug resistance rate and constituent ratio of various drug resistance patterns over the past seven years
Dynamic changes of the drug resistance rate of CHB patients with different genotypes over the past seven years
The changes in the drug resistance rate among CHB patients with different genotypes varied over the past seven years (Fig. 4a). From 2014 to 2020, the drug resistance rate of CHB patients with genotype B increased year by year, reaching the highest rate in 2020 (53.68%). From 2014 to 2020, the drug resistance rate of CHB patients with genotype C fluctuated from 40.16 to 54.89%, with the highest observed in 2015 (55.06%). The overall drug resistance rate among CHB patients showed an upward trend from 2014 to 2015. However, there was a subsequent decline from 2015 to 2016. From 2016 to 2020, the overall drug resistance rate resumed its upward trend, reaching a peak of 54.15% in 2020.
Dynamic changes in the drug resistance rate and various drug resistance patterns over the past seven years. (a) Dynamic changes in the drug resistance rate among CHB patients with different genotypes over the past seven years. Percentage = cases of genotype B(C) CHB patients with drug resistance per year/the total number of genotype B(C) CHB patients per year. (b) Dynamic changes in the constituent ratio of various drug resistance patterns over the past seven years. Percentage = cases of CHB patients with one drug resistance pattern per year/the total number of CHB patients with drug resistance per year; LAM: lamivudine; ETV: entecavir; ADV: adefovir; LDT: telbivudine.
Dynamic changes of the constituent ratio of various drug resistance patterns over the past seven years.
Meanwhile, the constituent ratio of various drug resistance patterns in CHB patients was distinct in different years (Fig. 4b). The most common single drug and multidrug resistance patterns were ADV and LAM + LDT, respectively. From 2014 to 2020, the proportion of ADV resistance ranged from 14.41 to 20.08%, showing an upward trend from 2014 to 2016, followed by a downward trend from 2016 to 2019, with the lowest proportion of 14.41% in 2019 and the highest proportion of 20.08% in 2016. From 2008 to 2014, the proportion of LAM + LDT resistance ranged from 69.04 to 73.48%, showing a relatively stable trend as a whole, with the lowest proportion of 69.04% in 2020 and the highest proportion of 73.48% in 2015.
Discussion
A hospital-based survey was conducted among 4266 chronic HBV patients in Southwest China to determine their HBV genotypes and assess drug resistance from January 2014 to December 2020. Genotypic resistance mutations were detected in 45.38% (1936/4266) CHB patients, of which 45.29% (1932/4266) were related to NAs resistance, higher than Italy (5%), Indonesia (13.5%) and Northern California (17.6%), lower than Korea (60.3%)27,34,35,36. Based on previous research, the proportion of drug-resistant mutations in Southwest China was higher than Northern (35.88%), Southern (16.34%), Eastern(30.78%) and Central China (8.92%)33,37,38,39. It is worth noting that the proportion of drug-resistant mutations in our study was lower than that in west of China (79.77%) conducted by Tao et al. in 201940. We guessed that it may be due to differences in the number of enrolled patients and ethnic groups included12. In our study, CHB patients in genotype B with genotypic mutations accounted for the highest proportion of all patients with genotypic mutations in 2019, accounting for 65.82%, which was consistent with the study by Tao et al. in 201940 .
As recommended in the Chinese Guidelines for Prevention and Treatment of CHB (2015 version), high genetic resistance barrier drugs should be preferred for CHB patients, and if low genetic resistance barrier drugs were used, the quality and combination therapy should be optimized. In addition, HBV genetic resistance test should be undertaken and appropriate adjustments to the treatment should be made accordingly in the case of treatment failure when HBV drug resistance is suspected41. The implementation of the new guideline effectively reduced the drug resistance rate of CHB patients in a short period of time. However, since 2016, resistance rates have gradually increased again, while new clinical challenges have been presented. The occurrence of low-level viremia (LLV) in patients with chronic HBV infection is associated with adverse outcomes, and it cannot be ruled out that the occurrence of LLV may be related to drug resistance of Entecavir and other nucleoside drugs.
Drug-resistant mutations to LAM was the most common site, consistent with the previous survey conducted in China42. Before 2012, LAM and ADV were the most widely used drug and primary choices for treatment due to their low genetic barrier to resistance. However, the Asian Pacific Association for the Study of the Liver (APASL) recommended ETV as the first choice in 2012 due to its low incidence of mutation site. Based on our survey, ETV exhibited a low mutation rate and a high genetic barrier to resistance, which suggested that ETV could be considered as the potential HBV antiviral drug. The ETV-resistant rate observed in our study was higher than the previous report, reaching 1.2% during the 5-year treatment period. This may suggest that the tendency of ETV resistance and long-term exposure to NAs could increase the risk of genotypic resistance, even for NAs with a high genetic barrier. Thus, more attention should be paid to the rising rate of ETV resistance in clinical practice.
Interestingly, the proportion of various drug resistance types in HBV patients changed with the passage of time. ADV and LAM + LDT are the most commonly observed single drug and multidrug resistance. The proportion of ADV resistance ranged from 14.41 to 20.08%, showing an upward trend from 2014 to 2016 and a subsequent downward trend from 2016 to 2019. From 2014 to 2020, the proportion of LAM + LDT resistance ranged from 69.04 to 73.48%, showing a relatively stable trend overall. The underlying reason for this phenomenon remained unclear, and a further long-term follow-up studies should be performed.
Many studies have investigated the impact of HBV genotypes on the proportion of drug resistance, but the correlation between drug resistance and mutation sites remained a controversial topic. One report found that HBV patients with genotype A had a higher LAM-resistance rate compared to those with genotype D43. While another study did not observe any significant differences in drug-resistant mutations between genotypes A and D44. Among genotypes B and C of HBV patients, rtM204I/V/S mutation (801,669) occurred more frequently, followed by rtL180M (286,343) and rtA181S/V/T (125,155), which was inconsistent with the previous study38. Multiple-site mutations rrtL180M + rtM204I/V (482/1936) were primarily detected in HBV genotypes C and B. Further research is needed to explore the underlying mechanisms.
There are several limitations in our study. Sanger sequencing only allows for the detection of one of the dominant virus strains, potentially overlooking the presence of mutant strains. The previous study demonstrated that next-generation sequencing (NGS) and high-throughput sequencing were more suitable than Sanger sequencing to monitor NAr mutations at a low rate in the treatment-naive patients and detect the resistance mutations quasispecies proportion of HBV DNA in NAs-treated CHB patients45,46. Furthermore, the treatment history and patients’ compliance were not clear. Nevertheless, the impact of antiviral treatment schedules and related biases could be mitigated by conducting investigations on a large sample size, and a comprehensive analysis of the evolution of drug-resistant HBV mutations could provide valuable insights for the effective treatment and prevention strategies.
The drug resistance mutation rate in HBV RT region increased in the Southwest of China from 2014 to 2020. LAM was the most common resistance drug, and rtM204I/V/S mutation was the most common mutation. More attention should be given to the increasing tendency of NAs resistance.
Materials and methods
Clinical information
A total of 4266 CHB patients who visited Qianjiang Hospital of Chongqing University between January 2014 and December 2020 were enrolled retrospectively. All enrolled CHB patients were characterized as HBsAg-positive ≥ six months, HBV DNA levels ≥ 1.0E + 04, absence of antibodies to HBsAg (anti-HBs), the persistent or intermittent elevation of ALT and (or) AST levels (> 40U/L), and with no evidence of cirrhosis or carcinoma by imaging and laboratory tests. Patients co-infected with hepatitis A, C, D, or human immunodeficiency virus (HIV) were excluded. All methods were carried out in accordance with relevant guidelines and regulations. This study was implemented after being approved by the Ethics Committee of Chongqing Qianjiang Central Hospital (Ethics Committee approval number: I2023213).
Instruments and reagents
The HBV-DNA extraction kit (Cat#B518223) was purchased from Sangon Biotech (Shanghai, China). The RT sequence of the HBV P region was amplified by polymerase chain reaction (PCR). The HBV-DNA sequencing kit BigDye™ Terminator v3.1 (Cat#4337455) was supplied by Applied Biosystems (ABI, USA). Test instruments included an ABI 7500 fluorescent PCR instrument and an ABI 3130 gene sequencer, manufactured by ABI.
Methods
HBV DNA extraction
The HBV DNA was extracted from the serum using HBV-DNA extraction kit from Sangon Biotech (Shanghai, China). In accordance to manufacturer’s instruction.
HBV DNA amplification
The primers for the P region of HBV DNA were the upstream primer (5′ -ACTGTATTCCCATCCCATCAT-3′) and the downstream primer (5′ -TTCGTTGACATACTTTCCAATCA-3′). The total PCR reaction system comprised of 2 µl of DNA template, 12.5 µl of PCR mix buffer, 2 µl of each upstream and downstream primer, and 8.5 µl of deionised water. The PCR cycle parameters were: holding at 42 °C for 5 min, pre-denaturation at 94 °C for 5 min, denaturation at 94 °C for 10 s, annealing at 55 °C for 30 s, extension at 72 °C for 40 s, 45 cycles, and constant temperature at 37 °C. The presence of PCR products was observed by 2% agarose gel electrophoresis.
Sequencing of PCR products after purification
Purified products were sequenced by ABI 3130. The sequencing results were then analyzed using NCBI BLAST and DNAMAN software to determine the mutation site and genotype.
Data analysis
The results of genotyping and drug-resistant mutation sites included in the analysis were analysed using SPSS 24.0 statistical software. And the counting data were compared between the groups using χ2 test. A p-value of < 0.05 was considered statistically significant.
Data availability
Due to China’s regulations on the management of human genetic resources, data sets generated and/or analyzed during the current study period are not publicly available, but are available from corresponding authors upon reasonable request.
References
Polaris Observatory C. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol.3 (6), 383–403 (2018).
Trepo, C., Chan, H. L. & Lok, A. Hepatitis B virus infection. Lancet. 384 (9959), 2053–2063 (2014).
Mortality, G. B. D. & Causes of Death, C. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the global burden of disease study 2013. Lancet. 385 (9963), 117–171 (2015).
Tsai, K. N., Kuo, C. F. & Ou, J. J. Mechanisms of hepatitis B virus persistence. Trends Microbiol.26 (1), 33–42 (2018).
McNaughton, A. L. et al. Insights from deep sequencing of the HBV genome-unique, tiny, and misunderstood. Gastroenterology. 156 (2), 384–399 (2019).
Fu, Y. et al. Mutational characterization of HBV reverse transcriptase gene and the genotype-phenotype correlation of antiviral resistance among Chinese chronic hepatitis B patients. Emerg. Microbes Infect.9 (1), 2381–2393 (2020).
Liu, T., Sun, Q., Gu, J., Cen, S. & Zhang, Q. Characterization of the tenofovir resistance-associated mutations in the hepatitis B virus isolates across genotypes a to D. Antiviral Res.203, 105348 (2022).
Ren, N. et al. Association of specific genotypes in metastatic suppressor HTPAP with tumor metastasis and clinical prognosis in hepatocellular carcinoma. Cancer Res.71 (9), 3278–3286 (2011).
Tao, J. et al. Prevalence of hepatitis B virus infection in Shenzhen, China, 2015–2018. Sci. Rep.9 (1), 13948 (2019).
Li, D. D. et al. Prevalence of hepatitis B virus genotypes and their relationship to clinical laboratory outcomes in Tibetan and Han Chinese. J. Int. Med. Res.38 (1), 195–201 (2010).
Kao, J. H. & Chen, D. S. HBV genotypes and outcome of HBV infection. Hepatology. 41 (1), 216 (2005). author reply 216.
Zhang, Q. et al. Epidemiology study of HBV genotypes and antiviral drug resistance in multi-ethnic regions from Western China. Sci. Rep.5, 17413 (2015).
Wiegand, J., Hasenclever, D. & Tillmann, H. L. Should treatment of hepatitis B depend on hepatitis B virus genotypes? A hypothesis generated from an explorative analysis of published evidence. Antivir Ther.13 (2), 211–220 (2008).
Watanabe, K. et al. Comparative study of genotype B and C hepatitis B virus-induced chronic hepatitis in relation to the basic core promoter and precore mutations. J. Gastroenterol. Hepatol.20 (3), 441–449 (2005).
Ni, Y. H. et al. Clinical relevance of hepatitis B virus genotype in children with chronic infection and hepatocellular carcinoma. Gastroenterology. 127 (6), 1733–1738 (2004).
Chu, C. J., Hussain, M. & Lok, A. S. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 122 (7), 1756–1762 (2002).
Watanabe, K. et al. Comparative study of genotype B and C hepatitis B virus-induced chronic hepatitis in relation to the basic core promoter and precore mutations. J. Gastroen Hepatol.20 (3), 441–449 (2005).
Cui, Y. & Jia, J. Update on epidemiology of hepatitis B and C in China. J. Gastroen Hepatol.28 (Suppl 1), 7–10 (2013).
Sunbul, M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J. Gastroenterol.20(18). (2014).
Lin, C. L. & Kao, J. H. The clinical implications of hepatitis B virus genotype: Recent advances. J. Gastroenterol. Hepatol.26 (s1), 123–130 (2011).
Cacciola, I. et al. Occult Hepatitis B virus infection in patients with chronic hepatitis C liver disease. New. Engl. J. Med.341 (1), 22–26 (1999).
Yu, M. W. et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: A prospective study in men. Jnci-J. Natl. Cancer I. 97 (4), 265–272 (2005).
Yu, M. W. et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: A prospective study in men. J. Natl. Cancer Inst.97 (4), 265–272 (2005).
Livingston, S. E. et al. Clearance of hepatitis B e antigen in patients with chronic hepatitis B and genotypes A, B, C, D, and F. Gastroenterology. 133 (5), 1452–1457 (2007).
Lin, C. L. & Kao, J. H. The clinical implications of hepatitis B virus genotype: Recent advances. J. Gastroenterol. Hepatol.26 (Suppl 1), 123–130 (2011).
Rajoriya, N., Combet, C., Zoulim, F. & Janssen, H. L. A. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J. Hepatol.67 (6), 1281–1297 (2017).
Mirandola, S. et al. Large-scale survey of naturally occurring HBV polymerase mutations associated with anti-HBV drug resistance in untreated patients with chronic hepatitis B. J. Viral Hepat.18 (7), e212–216 (2011).
Zoulim, F. & Locarnini, S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 137 (5), 1593–1608e1591 (2009).
Li, X. et al. Comparison of detection rate and mutational pattern of drug-resistant mutations between a large cohort of genotype B and genotype C hepatitis B virus-infected patients in North China. Microb. Drug Resist.23 (4), 516–522 (2017).
Qian, F. et al. Monitoring of genotypic resistance profile in chronic hepatitis B patients receiving nucleos(t)ide analogues in Huzhou, China. J. Infect. Dev. Ctries.10 (9), 996–1002 (2016).
Meng, T. et al. Analysis of the prevalence of drug-resistant hepatitis B virus in patients with antiviral therapy failure in a Chinese tertiary referral liver centre (2010–2014). J. Glob. Antimicrob. Resist.8, 74–81 (2017).
Liu, Y. et al. Genotypic resistance profile of hepatitis B virus (HBV) in a large cohort of nucleos(t)ide analogue-experienced Chinese patients with chronic HBV infection. J. Viral Hepat.18 (4), e29–39 (2011).
Guo, X. et al. Trends in hepatitis B virus resistance to nucleoside/nucleotide analogues in North China from 2009–2016: A retrospective study. Int. J. Antimicrob. Agents. 52 (2), 201–209 (2018).
Kim, J. E. et al. Naturally occurring mutations in the reverse transcriptase region of hepatitis B virus polymerase from treatment-naïve Korean patients infected with genotype C2. World J. Gastroenterol.23 (23), 4222–4232 (2017).
Yamani, L. N. et al. Profile of mutations in the reverse transcriptase and overlapping surface genes of hepatitis B virus (HBV) in treatment-Naïve Indonesian HBV carriers. Jpn J. Infect. Dis.70 (6), 647–655 (2017).
Nguyen, M. H. et al. Prevalence of hepatitis B virus DNA polymerase mutations in treatment-naïve patients with chronic hepatitis B. Aliment Pharmacol. Ther.30 (11–12), 1150–1158 (2009).
Chen, X. H., GD, X. & SQ, L. Z. N: Study on hepatitis B virus genotype and drug resistance gene mutation in chronic hepatitis B patients in Guangxi, Yunnan and Guizhou border areas. Vaccines and Immunization in China 27(4)366–369 2021. (2021).
Zhang, H. Y. et al. Evolution of drug-resistant mutations in HBV genomes in patients with treatment failure during the past seven years (2010–2016). Virus Genes. 54 (1), 41–47 (2018).
Zhao, Y. et al. Prevalence of mutations in HBV DNA polymerase gene associated with nucleos(t)ide resistance in treatment-naive patients with chronic hepatitis B in Central China. Braz J. Infect. Dis.20 (2), 173–178 (2016).
Tao, X. et al. Genotyping and drug resistance mutation analysis of hepatitis B virus in West China Hospital of Sichuan University. West. China Med.2019(8):5 .
Hou, J. et al. Guideline of prevention and treatment for chronic hepatitis B (2015 update). J. Clin. Transl Hepatol.5 (4), 297–318 (2017).
Lee, A. J., Lee, C. H. & Jeon, C. H. Analysis of reverse transcriptase gene mutations in the hepatitis B virus at a university hospital in Korea. Ann. Lab. Med.34 (3), 230–234 (2014).
Zollner, B. et al. Viral features of lamivudine resistant hepatitis B genotypes a and D. Hepatology. 39 (1), 42–50 (2004).
Buti, M. et al. Is hepatitis B virus subtype testing useful in predicting virological response and resistance to lamivudine? J. Hepatol.36 (3), 445–446 (2002).
Fu, Y. et al. Characterization and clinical significance of natural variability in hepatitis B virus reverse transcriptase in treatment-Naive Chinese patients by Sanger sequencing and next-generation sequencing. J. Clin. Microbiol.57(8). (2019).
Liao, H. et al. Nucleos(t)ide analogues altered quasispecies composition of hepatitis B virus (HBV)-resistant mutations in serum HBV DNA and serum HBV RNA. J. Med. Virol.95 (3), e28612 (2023).
Funding
This work was funded by the Chongqing medical scientific research project (Joint project of Chongqing Health Commission and Science and Technology Bureau) (2024MSXM008 to C-L.H.), the 2024 Chongqing Young and Middle-aged High-level Medical Talent Project of C-L.H., the Postdoctoral research project of Chongqing Natural Science Foundation (CSTB2023NSCQ-BHX0235 to C-L.H.), the China Postdoctoral Science Foundation (2023M740443 to C-L.H.), the Chongqing postdoctoral research project special support (2023CQBSHTB3076 to C-L.H.), the Fundamental Research Funds for the Central Universities (2023CDJYGRH-YB16 to Y-H.W.), the National Natural Science Foundation of China (82171440 to X.X.), the Chongqing Key Clinical Specialized Funding, the Chongqing Natural Science Foundation project (CSTB2022NSCQ-MSX0892 to M-J.Z.), the Chongqing Natural Science Foundation project (CSTB2022NSCQ-MSX0221 to Z-L.Y.) and Chongqing Health appropriate technology extension project (2023jstg047 to Z-L.Y).
Author information
Authors and Affiliations
Contributions
Y.W., C.H., M.Z., X.W., Z.Y., L.D., and X.X. designed the research. L.W., X.Y., Z.X., X.Z., Z.C., Q.L., T.Z., and H.H. collected and curated the Data. C.H. and Z.Y. analyzed and verified the data. Y.W., X.W., and L.D. interpreted the results and wrote the first draft. X.X. and M.Z. contributed to the manuscript revision. Z.T. sorted out the clinical treatment strategies and outcomes of the cases, and discussed the reasons for the changes in the revised manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Transparency declarations
None to declare.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, C., Wu, X., You, Z. et al. Evaluation of genotype characteristics and drug resistance mutations in patients with chronic hepatitis B. Sci Rep 14, 27575 (2024). https://doi.org/10.1038/s41598-024-77362-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-77362-1
Keywords
This article is cited by
-
Molecular epidemiology of hepatitis B virus from blood samples obtained from persons in Enugu and Nasarawa States, Nigeria
BMC Infectious Diseases (2025)
-
Progress and gaps in antimicrobial resistance research within One Health sectors in China: a systematic analysis
Science China Life Sciences (2025)