Abstract
The COVID-19 pandemic highlighted the need for the rapid development of antiviral therapies. Viral RNA-dependent RNA polymerases (RdRp) are promising targets, and numerous virtual screenings for potential inhibitors were conducted without validation of the identified hits. Here we have tested a set of presumed RdRp inhibitors in biochemical assays based on fluorometric detection of RdRp activity or on the electrophoretic separation or RdRp products. We find that fluorometric detection of RdRp activity is unreliable as a screening method because many small compounds interfere with fluorophore binding to dsRNA, and this effect is enhanced by the Mg2+ metal ions used by nucleic acid polymerases. The fact that fluorimetric detection of RdRp activity leads to false-positive hits underscores the requirement for independent validation methods. We also show that suramin, one of the proposed RdRp inhibitors that could be validated biochemically, is a multi-polymerase inhibitor. While this does not hinder its potential as an antiviral agent, it cannot be considered an specific inhibitor of SARS-CoV-2 RdRp.
Similar content being viewed by others
Introduction
The COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the rapid development of RNA-based vaccines and highlighted the need for efficient antiviral therapies to alleviate the clinical consequences of new viral outbreaks. When the efficacy of vaccines is compromised due to the high mutagenic rate of the virus or its evasion of the immune system, antiviral drugs become the main tool to fight emerging diseases.
Virus-encoded DNA or RNA polymerases are sufficiently different from their human counterparts to make them targets for antiviral drug development. Successful examples in the clinic include polymerase inhibitors for herpesvirus (acyclovir), cytomegalovirus (foscarnet), hepatitis C virus (sofosbuvir, ribavirine), hepatitis B virus (tenofovir, lamivudine) and the HIV reverse transcriptase (efavirenz, rilpivarine, ritonavir, etravirine, nevirapine). The genome of coronaviruses consists of a single-stranded RNA molecule that requires an RNA-dependent RNA-polymerase (RdRp) for replication and transcription. SARS-CoV-2 RdRp is formed by non-structural proteins (nsp)12, nsp7 and nsp8. High-resolution structures of nsp12-nsp7-nsp8 generated by cryo-electron microscopy1,2,3 have served as the basis for many virtual screening campaings searching for potential inhibitors based on docking simulations4,5,6,7. In contrast with these in silico approaches, only a handful of functional screenings have been reported using fluorometric8,9, strand-displacement10 or cell-based reporter assays11,12,13.
The biochemical activity of DNA or RNA polymerases can be analyzed in vitro using primer extension assays followed by electrophoretic separation of the extension products. However, this classical approach is impractical for the high-throughput (HTP) screening of large libraries of small compounds. One technical alternative is the use of fluorophores such as SYBRGreen, PicoGreen, or Quantifluor dsDNA, which drastically enhance their basal fluorescence upon interaction with double-stranded DNA (dsDNA) or double-stranded RNA (dsRNA). The fluorescent emission of these agents bound to newly synthesized dsDNA or dsRNA is used to quantify the polymerase reaction in HTP screenings. Fluorometric methods were originally applied to enzymes capable of de novo DNA/RNA synthesis14,15,16,17, but polymerases lacking primase activity can also be monitored using template oligonucleotides with a short self-priming section8,9,18. These assays have identified RdRp inhibitors of Hepatitis C virus14, Zika virus15,16, West Nile virus17 and more recently SARS-CoV-28,9. In addition, they have been used as an alternative to electrophoretic methods to quantify polymerase activity19,20.
In this study, we have tested a set of potential SARS-CoV-2 RdRp inhibitors selected from published studies in fluorometric and electrophoretic RdRp polymerization assays, revealing key technical concerns about nucleic acid intercalant-based fluorometric HTP screenings.
Results
Putative RdRp inhibitor compounds interfere with fluorophore-based activity assays
SARS-CoV-2 nsp12-7-8 proteins, which form the functional RdRp, were co-expressed in E. coli and purified as described21 (Fig. 1A). A selection of 15 putative RdRp inhibitors were chosen for a detailed analysis of their activity, based on several criteria. First, we included three molecules for which some functional validation has been reported (suramin, corilagin and simeprevir). From the published virtual screening studies, we included commercially available compounds predicted to be RdRp inhibitors by more than one study (baloxavir marboxil, elbasvir, fidaxomicin, fluvastatin, grazoprevir, ivermectin, paritaprevir, rifabutin, rifampicin, verbascoside), and compounds that were readily available at the CNIO Experimental Therapeutics Program drug repository (rifapentine, ruxolitinib). We noticed that some of these compounds are antibiotics with nucleic acid polymerase inhibitory capacity (rifampicin, rifapentine, fidaxomicin). Two FDA-approved small molecules not reported as putative RdRp inhibitors (sildenafil and duvelisib) were added as controls. For the full list of compounds, see Supplementary Table 1.
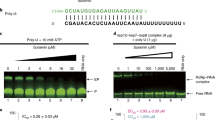
Putative RdRp inhibitors display different activity in fluorophore-dependent and fluorophore-independent assays. A Left, schematic of the purification procedure of the nsp12-nsp8-nsp7 complex comprising the functional SARS-CoV-2 RdRp. Right, Coomassie Blue staining following electrophoretic separation of the purified RdRp complex. The positions of the three subunits are indicated. Different amounts of bovine serum albumin (BSA) are shown and were used to estimate nsp12 concentration. B Top, schematic of the fluorophore (SYBR Green)-dependent detection of RdRp activity. Bottom, dose-response assay with purified recombinant SARS-CoV-2 RdRp. C Histogram showing the inhibitory effect of the 17 test compounds as assessed in a SYBR Green-based assay. D Top, schematic of the electrophoretic-based detection of RdRp polymerization activity. Bottom, dose-response assay with purified SARS-CoV-2 RdRp. E Inhibitory activity of the 17 test compounds on RdRp activity using the electrophoretic method. For quantification purposes, the signal contained in areas A and B (see control lane) was measured with ImageJ, and polymerization efficiency (PE) was estimated by the formula PE = A/(A + B). The PE value in the absence of inhibitors (control lane) was approx. 68–70%. Histogram represents the percentage of polymerization in the presence of each inhibitor, relative to the control value (plotted as 100%). The quantification includes data from two independent electrophoretic experiments. Original gel images for parts A, D and E are presented in Supplementary Fig. 3.
RdRp activity was first tested in a fluorometric assay using a self-priming RNA molecule consisting of a 10 bp section of dsRNA followed by a 30 nt-long poly(U)18 sequence. The binding of the SYBR Green fluorophore to newly synthesized dsRNA allowed the indirect quantification of RNA polymerase activity (Fig. 1B). Notably, only 8 of the 15 putative inhibitors decreased the fluorescent signal to less than 60% of the control reaction, while the rest displayed modest or no inhibition. The highest RdRp inhibitory potential was observed with suramin, rifapentin, verbascoside, corilagin and simeprevir, while the two control molecules displayed no inhibition (Fig. 1C). In parallel, RdRp activity was tested by primer-extension of a short 5’-fluorescently labeled RNA primer hybridized to a longer RNA template. In this assay, extension products are separated electrophoretically eliminating the need for SYBR Green (Fig. 1D). Strikingly, most compounds displayed little or no inhibition of RdRp primer-extension activity (Fig. 1E), revealing that the fluorometric detection of RdRp activity is prone to identifying false-positive hits. Only corilagin, suramin and simeprevir, previously validated RdRp inhibitors, and to a lesser extent verbascoside, displayed inhibitory activity in this assay. Combined, these observations denote a high rate of false-positive hits in fluorophore-based assays, at least in the experimental conditions used in our study.
Interference of putative SARS-CoV-2 RdRp inhibitors with SYBR Green binding to RNA
The apparent inhibition of RdRp activity by several compounds in the fluorometric polymerase assay shown in Fig. 1B could be indirectly caused by interference with the dsRNA-binding fluorophore. To test this possibility, we performed fluorescence displacement assays22,23,24,25 that monitor the capacity of compounds to interfere with the fluorescence emission of nuclei acid-binding fluorophores such as SYBR Green. Interference could be caused by different reasons: (i) displacement of fluorophores from the nucleic acid; (ii) direct interaction with the fluorophore (quenching); (iii) alterations of nucleic acid structure that decrease the affinity for fluorophores (Fig. 2A). Strikingly, at 50 µM concentration, 11 of the 15 compounds (including suramin, corilagin and simeprevir) strongly interfered with the fluorescent emission of SYBR Green bound to poly(AU)15 dsRNA (Fig. 2B). In contrast, the majority of these compounds did not have any impact on SYBR Green fluorescence on dsDNA (Fig. 2C). These results suggest a more labile association of SYBR Green with dsRNA than with dsDNA.
SYBR Green fluorescence displacement assays from dsRNA and dsDNA. A Schematic of the SYBR Green displacement assay from double-stranded nucleic acid molecules. B Histogram shows the SYBR Green displacement from a poly-(AU)15 dsRNA by each of the 17 test compounds. C Same as (B), testing SYBR Green displacement from a poly-(AT)15 DNA molecule. See text for details.
Mg2+ affects the association of fluorophores with nucleic acids
We noticed that the presence of Mg2+, the metal cofactor used by RdRp during polymerization reactions, exacerbated the displacement of SYBR Green on dsRNA caused by suramin, corilagin and simeprevir (Fig. 3A). Interestingly, displacement of SYBR Green from dsDNA required a 10-fold higher concentration of suramin than SYBR Green displacement from dsRNA, and was not observed with corilagin and simeprevir (Fig. 3B). Mg2+ reduced the fluorescence of other fluorophores such as PicoGreen and Quantifluor on dsRNA to a much higher extent than on dsDNA (Fig. 3C). PicoGreen and Quantifluor were displaced by suramin from dsRNA and dsDNA, recapitulating the results obtained with SYBR Green (Supplementary Fig. 1). Combined, these results indicate that fluorophore-dsRNA fluorescence is easily perturbed by small molecules, particularly in the presence of Mg2+, and this effect compromises the identification of RdRp inhibitors by fluorometric methods.
Mg2+affects the association of fluorophores with nucleic acids. A Dose-response activity of suramin, corilagin, and simeprevir on the fluorophore displacement of SYBR Green from poly-(AU)15 dsRNA (in the presence or absence of 6 mM MgCl2. B Same as (A), using poly-(AT)15 DNA. C Effect of MgCl2 concentration on the fluorescent emission of SYBR Green, PicoGreen, and Quantifluor DNA bound to dsRNA (blue) or dsDNA (green).
Suramin is an efficient but non-specific SARS-CoV-2 RdRp inhibitor
Suramin is the only putative inhibitor whose binding to RdRp has been reported by cryo-EM20 and surface plasmon resonance26. Dose-response assays indicated that suramin was a more efficient inhibitor of RdRp than corilagin and simeprevir (Fig. 4A). However, besides its clear inhibitory effect in electrophoretic-based RdRp assays (Fig. 1E), it also displaced SYBR Green fluorescence from dsRNA and dsDNA (Figs. 2 and 3), causing misleading readouts in fluorometric polymerase assays. Suramin is a polyanionic compound and its interaction with nucleic acids is unlikely, but it might interact directly with SYBR Green. Consistent with this hypothesis, suramin increased SYBR Green auto-fluorescence (in the absence of nucleic acids) in a dose-dependent manner (Supplementary Fig. 2A). SYBR Green increases its fluorescence when bound to dsDNA as its intra-molecular motions are reduced27. Similarly, the binding of suramin may estabilize the SYBR Green molecule in solution, increasing its fluorescence and preventing its association with dsRNA/dsDNA. It is relevant to mention that in the fluorophore-based RdRp assays, the increase in SYBR Green fluorescence caused by suramin was not observed because the fluorophore molecule was used at much smaller concentration.
We next assessed the specificity of suramin by testing its effect on other polymerases, i.e. Φ29 bacteriophage DNA polymerase (Φ29pol), human mitochondrial DNA polymerase (Polγ) and human Primase and DNA-directed Polymerase (PrimPol). Strikingly, suramin inhibited the activity of all three enzymes (Fig. 4B). Because suramin contains six negatively charged sulfonate groups, its interaction with polymerases could be mediated by electrostatic interactions with the positive-charged pockets present in DNA-binding proteins. In agreement with this notion, it has been reported that suramin prevents the binding of SARS-CoV-2 RdRp to RNA20,26. Besides, we have observed that suramin prevents the association of Φ29pol to DNA (Supplementary Fig. 2B). In addition, suramin inhibited the activity of other DNA-binding enzymes such as endonucleases EcoRI, XbaI, KpnI, ApaI and NotI (Fig. 4C). Suramin did not inhibit other restriction enzymes such as BamHI, XhoI, and EcoRV, suggesting that its interaction with target proteins is also restricted by conformational constraints.
Suramin is a multi-polymerase inhibitor. A Dose-response assays showing the RdRp inhibitory activity of suramin, corilagin, and simeprevir in the electrophoretic-based polymerase assay. B Dose-response effect of suramin on the activity of Φ29pol, Polγ, and PrimPol. C Left, schematic of pCDNA3 BglII-XmaI fragment (2,078 bp) depicting the restriction enzymes whose target sequences are present in the polylinker region. Right, agarose gel electrophoretic analysis of the pCDNA3 BglII-XmaI fragment following incubation with the indicated restriction enzymes in the presence or absence of suramin (200 µM). Original gel images are presented in Supplementary Fig. 3.
Taken together, these observations indicate that suramin is a non-specific inhibitor of viral and eukaryotic DNA and RNA polymerases that can also inhibit other nucleic acid-binding enzymes. This promiscuity is likely explained by electrostatic interactions that interfere with the binding of proteins to dsDNA.
Discussion
The COVID-19 pandemic triggered the search for inhibitors of RdRp, the enzyme responsible for viral genome replication28. HTP screenings for RdRp inhibitors may rely on the indirect quantification of polymerization products using nucleic-acid binding fluorophores, such as SYBR Green8,9,10,14,15,16,17,18. While fluorescence-based assays are effective tools for small molecule screening campaigns, they are also limited by the potential interference of compounds with the fluorescent signal, either by autofluorescence, fluorophore quenching or interference with fluorophore binding to nucleic acids29,30. In this study, we have reported that the fluorescence emission of SYBR Green bound to dsRNA is compromised in the presence of many small compounds, imposing a critical technical limitation for the fluorophore-based identification of RdRp inhibitors. This limitation arises from the low affinity of nucleic-acid binding fluorophores for dsRNA, which is exacerbated by the presence of Mg2+ ions that act as cofactors in the polymerization reaction (see model in Fig. 5). In contrast, fluorescent dyes exhibit a robust interaction with dsDNA that is more resistant to interference by other molecules. The inherent differences in composition and helical structures between dsRNA and dsDNA likely account for their varying affinity for fluorophores and the differential influence of the ionic environment31,32. In fluorescence displacement assays, approximately 65% of the small molecules tested interfered with the binding of SYBR Green to dsRNA. It is worth noting that fluorescent displacement does not automatically rule out RdRp inhibition capacity, as exemplified by suramin that exhibited strong fluorescence displacement and also inhibited RdRp in SYBR Green-independent assays (this study and published data10,20).
Out of 15 potential SARS-CoV-2 RdRp inhibitors tested, only suramin, corilagin and simeprevir displayed marked inhibition of RdRp in SYBR Green-independent RNA polymerase reactions. Suramin, a trypanocidal drug used to treat sleeping sickness33, exhibited the stronger activity. Corilagin, a plant polyphenol with antioxidant properties34, and simeprevir, used to target the hepatitis C virus (HCV) protease35,36, showed comparatively weaker inhibitory effects. Other putative RdRp inhibitors could not be validated biochemically. We infer from these data that the success of virtual screenings strongly depends on the availability of accurate structural data and thorough post-validation through molecular dynamics simulations or enhanced sampling techniques37,38 followed by independent biochemical validation to filter out false positives.
Suramin inhibited other DNA-binding enzymes including DNA polymerases, primases, and restriction enzymes. This promiscuous activity is likely due to its high degree of conformational flexibility39 and its six negative charges at physiological pH, which facilitate electrostatic interactions with the positively charged protein domains that typically mediate association with nucleic acids. Indeed, nucleic acid-binding proteins are common targets of suramin30,34, and its inhibitory activity could arise from preventing their interaction with dsDNA or dsRNA. In this regard, pioneering studies published over 35 years ago first reported the ability of suramin to target DNA and RNA polymerases40,41,42. Still, the list of suramin targets goes beyond nucleic acid-binding proteins. In the context of SARS-CoV-2, suramin has been reported to interact with the 3CL protease43,44, the nucleocapsid protein43,44, the Spike glycoprotein45,46,47 and the nsp13 helicase48. This broad-spectrum of inhibitory activity complicates the attribution of its antiviral effects against SARS-CoV-2 infection in cellular systems20,46,49 to a single molecular mechanism. In line with this, suramin exhibits a broad-spectrum antiviral activity, encompassing duck hepatitis B virus50, Hepatitis C virus51, Rous Sarcoma virus50, Chikungunya virus52, Ebola virus48,52,53, Zika virus54,55, EV7156 and Mayaro viruses57. Despite logical concerns about its multi-target effects, potential toxicity and limited bioavailability33,58, suramin has been used for more than 100 years to treat human African Trypanosomiasis (sleeping sickness)33, suggesting that its clinical usefulness should not be disregarded, as it may serve as a first line of defense against emerging viruses before more specific therapeutic agents are developed. However, its efficacy against COVID-19 remains to be proven.
Methods
Expression and purification of SARS-CoV-2 RdRp complex
The bacterial expression system for RdRp purification from pRSFDuet-1(nsp8/nsp7)(nsp12) plasmid21 was kindly provided by Dr. L. Sauguet (Unit of Structural Dynamics of Macromolecules, Institut Pasteur & CNRS UMR, Paris, France). The RdRp complex was expressed and purified essentially as reported21 using the following protocol: E. coli BL21 Star (DE3) strain (ThermoFisher) transformed with pRSFDuet-1(nsp8/nsp7)(nsp12) was grown at 37 oC in the presence of 100 µg/ml kanamycin until the optical density at 600 nm (OD600) reached 0.6. Protein expression was induced with 0.05 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 20 °C for 20 h. Cells were harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at −80 °C. The bacterial pellet was resuspended in HisTrap buffer A (50 mM HEPES, pH 7.5, 500 mM NaCl, 10 mM imidazole) supplemented with EDTA-free protease inhibitors (Roche) and 500 units of benzonase (Sigma). The cells were lysed by sonication (5 min; 1 s on/ 1 s off) using a Vibra-Cell™ 75,042 ultrasonic cell disruptor at 40% amplitude, and the lysate was cleared by centrifugation (14,000 rpm/ 30 min/ 4 oC). The cleared supernatant was incubated with nickel affinity agarose beads (ABT) for 2 h. The beads where packed in a chromatography column, washed with HisTrap buffer A + 5% HisTrap buffer B (50 mM HEPES, pH 7.5, 500 mM NaCl, 500 mM imidazole), and eluted in 1 ml fractions with 5 ml of 50% HisTrap buffer A + 50% HisTrap buffer B. The next steps were performed on a AKTA pure HPLC system (GE Healthcare). The HisTrap fractions containing the eluted proteins were pooled, diluted 5-fold with 50 mM HEPES, pH 7.5 and loaded in a HiTrap Q anion exchange column (GE Healthcare) pre-equilibrated with HiTrap Q buffer A (50 mM HEPES, pH 7.5, 150 mM NaCl). The column was washed with HiTrap Q buffer A and eluted in a linear NaCl gradient formed by gradually mixing HiTrap Q buffer A and HiTrap Q buffer B (50 mM HEPES, pH 7.5, 1 M NaCl). The fractions containing the RdRp complex were pooled and concentrated using an Amicon Ultra-15-centrifugal filter unit MWCO 10 kD (Millipore). Finally, the complex was loaded on a Superdex 200 column equilibrated with S200 buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 1mM MgCl2) and purified by size-exclusion chromatography. The fractions containing the RdRp proteins were combined, concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C. The concentration of the catalytic subunit (nsp12), estimated by comparison with reference concentrations of bovine serum albumin (BSA), was ~ 0.5 µM.
Expression and purification of PrimPol and other polymerases
The pET16-10His-PrimPol plasmid containing the histidine-tagged cDNA encoding for PrimPol59 was kindly provided by Dr. L. Blanco (CBM Severo Ochoa, CSIC-UAM, Madrid). The plasmid was transformed into the E. coli BL21 (DE3) pRIL strain, and the bacteria were grown in the presence of ampicillin (100 µg/ml) and chloramphenicol (15 µg/ml) at 30 °C until the OD600 reached 0.6. Protein expression was induced with 0.5 mM IPTG at 30 °C for 3 h. The cells were harvested by centrifugation and resuspended in Lysis Buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, 10% glycerol, 15 mM imidazole) supplemented with 2 mM β−mercaptoethanol, 1 mM PMSF, and EDTA-free protease-inhibitor cocktail (Roche). The bacterial suspension was then lysed by sonication as described above. The lysate was cleared by centrifugation at (14,000 rpm/ 30 min). The cleared supernatant was incubated with nickel affinity agarose beads (ABT) for 2 h and loaded into a chromatography column. The column was washed with Lysis Buffer and eluted with Lysis Buffer + 300 mM Imidazole. The eluted fractions containing PrimPol were pooled, diluted 5-fold with HiTrap Heparin buffer A (50 mM Tris-HCl, pH 8, 500 mM NaCl, 10% glycerol) and loaded into a HiTrap Heparin HP column (GE Healthcare) connected to an AKTA pure HPLC system (GE Healthcare). The column was washed with HiTrap Heparin buffer A and eluted with a linear NaCl gradient formed by gradually mixing HiTrap Heparin buffer A and HiTrap Heparin buffer B (50 mM Tris-HCl, pH 8, 1 M NaCl, 10% glycerol). The fractions containing PrimPol were pooled, glycerol was added to a final concentration of 50%, and the samples were flash-frozen in liquid nitrogen and stored at -80 °C.
Polγ was kindly provided by Dr. Araceli Grande-García (Genomic Integrity and Structural Biology department, CNIO, Madrid, Spain). Exonuclease-deficient Φ29pol was kindly provided by Dr. Miguel de Vega (CBM Severo Ochoa, CSIC-UAM, Madrid, Spain).
Fluorometric polymerase assay
This method was adapted from a protocol initially developed for detecting of Hepatitis C virus RNA polymerase activity14 and subsequently applied to identify inhibitors of Zika virus15,16, West Nile virus17 and, more recently, SARS-CoV-2 virus8,9,60. The reactions were performed in duplicates using 384-well flat-bottom polystyrene plates (Corning 3575). The self-priming RNA template (5´-U30AACAGGUUCUAGAACCUGUU-3´) was previously described18.
The standard reaction was conducted in 20 µl containing 25 mM Tris-HCl, pH 8, 6 mM MgCl2, 200 nM self-priming oligo, 15 µM ATP and 1:20,000 SYBR Green (Sigma). The reaction was initiated by adding the SARS-CoV-2 RdRp complex and carried out at 30 ºC for 60 min. The estimated final concentration of nsp12 was 25 nM. Fluorescence was measured using a Victor3 1420 Multilabel Counter (Perkin Elmer) with excitation and emission filters set at 497 nm and 520 nm, respectively.
The setup for the inhibition assays was identical, except compounds were pre-incubated with the RdRp complex for 10 min before initiating the reaction with the remaining components. Reactions were conducted in the presence of 0.01% Triton X-100 to prevent compound aggregation.
Electrophoretic-based assay of SARS-CoV-2 RdRp polymerase activity
A fluorescein-labeled ribo-oligonucleotide (5´-FAM-GCUAUGUGAGAUUAAGUUAU-3´) was used as primer, and a longer RNA oligonucleotide (5´-U10AUAACUUAAUCUCACAUAGC-3´) (IDT) was used as template as described28. Oligonucleotides were from Integrated DNA Technologies (IDT). Both RNA oligonucleotides were mixed in Annealing Buffer (10 mM Tris-HCl, pH 8, 25 mM NaCl and 2.5 mM EDTA), heated to 90 C for 5 min and slowly cooled to room temperature. Primer extension assays were performed in 10 µl for 1 h at 30 C. The reaction mixture contained 25 mM Tris-HCl, pH 8, 6 mM MgCl2, 90 nM RNA duplex and 40 µM ATP. Reactions were initiated by adding 1 µl of purified complex. The estimated final concentration of nsp12 was 50 nM. The reactions were stopped with 10 µl of formamide loading buffer (5 mM EDTA, 0.025% w/v Xylene Cyanol, 0.025% w/v bromophenol blue, 95% formamide) and heating 5 min at 90 C. Reaction products were resolved in 10% urea-PAGE gels (SequaGel) and imaged on a Typhoon TRIO (GE Healthcare).
Inhibition assays were identical except that compounds were pre-incubated with RdRp complex for 10 min before initiating the reaction with the remaining components. Reactions were performed in the presence of 0.01% Triton X-100 to prevent compound aggregation.
Electrophoretic-based assay of PrimPol, Φ29pol and Polγ polymerase activity
For PrimPol, Φ29pol (exonuclease-deficient) and Polγ, a short fluorescein-labeled DNA oligonucleotide (5´FAM-TCCGAGCGAGCGAGCGAGCCT-3´) was used as primer after being annealed to the oligo template (5´-T12AGGCTCGCTCGCTC GCTCGGA-3´) (IDT) as previously described. PrimPol reaction contained 50 mM Tris-HCl pH, 7.5, 1 mM MnCl2, 40 µM dATP and 200 nM template-primer duplex, and 120 nM PrimPol. For Φ29pol, the conditions were identical except that the reaction included 6 mM MgCl2 instead of 1 mM MnCl2. Polγ reactions were performed in 50 mM Tris-HCl, pH 7.5 containing 30 mM KCl, 2 mM DTT, 4 mM MgSO4, 0.1 mg/µl BSA, 10% glycerol, 40 µM dATP, and 200 nM template-primer and 100 nM enzyme.
Fluorescence displacement assay
The DNA oligonucleotides 5´-AAAAAAAAAAAAAAA-3´ (poly-dA)15 and 5´-TTTTTTTTTTTTTTT-3´(poly-dT)15 (Invitrogen), and the RNA oligonucleotides poly-(A)15 and poly-(U)15 were annealed as described above. The resulting DNA (poly-AT)15 and RNA (poly-AU)15 duplexes were incubated with either 1:20,000 SYBR Green (Sigma), 1:2000 Quantifluor dsDNA (Promega) or 1:400 PicoGreen (Life Technologies) in the presence or absence of MgCl2 and the indicated concentrations of compounds.
The assays were performed in duplicates in 384-well plates flat bottom polystyrene plates (Corning 3575) as follows: 10 µl of compound diluted in 25 mM Tris-HCl, pH 8 plus 0.02% Triton X-100 were added to 10 µl of 25 mM Tris-HCl, pH 8 with either 0.8 µM of (poly-AT)15 or 1.6 µM of (poly-AU)15 and the corresponding fluorophore. When indicated, 6 mM MgCl2 was included in the mix. Fluorescence was recorded 10 min after addition of the compounds to the nucleic acid and fluorophore, using a Victor-3 1420 Multilabel counter (Perkin Elmer) with excitation and emission filters set at 497 nm and 520 nm, respectively. The effects of each compound on dsRNA and dsDNA were normalized to control conditions using DMSO.
Electrophoretic mobility shift assay (EMSA)
Exonuclease-deficient Φ29pol enzyme (0.6 µg) was incubated with 4.4 µM fluorescein-labeled DNA template/primer in the presence of absence of the indicated concentrations of suramin in 25 mM Tris-HCl, pH 8 buffer containing 6 mM MgCl2 and 0.01% Triton X-100. Following a 20 min incubation at RT, the samples were resolved in non-denaturing 1% acrylamide gels. Oligonucleotides: 5´- TTTTTTTTTTTTAGGCTCGCTCGCTCGCTCGGA-3´ and 5´FAM-TCCGAGCGAGCGAGCGAGCCT-3´ were annealed as described prior to incubation with the polymerase.
Effect of suramin on DNA endonuclease digestions
A 2,078 bp DNA fragment of pCDNA3.1 plasmid containing the polylinker region was generated by digestion with XmaI and BglII enzymes. 50 ng of the purified DNA fragment were incubated with the indicated restriction enzymers (New England Biolabs) in the presence or absence of 200 µM suramin by 1 h at 37 C in the manufacturer recommended buffers. Products were analyzed by agarose electrophoresis.
Equipment and settings
Fluorometric assays of RdRp activity and fluorescence displacement assays (Figs. 1B-C, 2B-C and 3A-C, Supplementary Fig. 1A-B and Supplementary Fig. 2A): fluorescence was measured in a Victor-3 1420 Multilabel Counter (Perkin Elmer) with excitation and emission filters set at 497 and 520 nm, respectively. Electrophoretic RdRp assays (Figs. 1D-E and 4A-B) and the band-shift assay (Supplementary Fig. 2B) gels were imaged in a Typhoon TRIO (GE Healthcare). Quantification of Fig. 1E was done with ImageJ.
Restriction digestion assays (Fig. 4C) were analysed by agarose gel electrophoresis with SYBR-Safe (Invitrogen) and imaged in a Molecular Imager Gel Doc XR System (Bio-Rad).
Statistical analysis and model figures
All SYBR Green-based assays were performed at least three times, with each experiment containing two technical replicates. Results are expressed as mean and standard deviation. Dose-response fluorometric assays were fitted to sigmoidal curves using GraphPad Prism software with nonlinear regression. IC50 values were obtained by fitting the data to a four-parameter logistic equation. Models and schematics were created with BioRender.
Chemical compounds
The compounds were obtained from the indicated companies (Supplementary Table 1). 10 mM stocks were prepared in DMSO.
Data availability
Data used to generate the results in this study is provided within the manuscript and supplementary information files. All raw data is available upon reasonable request to any of the corresponding authors: S.L. and J.M.
References
Kirchdoerfer, R. N. & Ward, A. B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 (2019).
Hillen, H. S. et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 584, 154–156 (2020).
Gao, Y. et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 368, 779–782 (2020).
Bajad, N. G. et al. Systematic review on role of structure based drug design (SBDD) in the identification of anti-viral leads against SARS-Cov-2. Curr. Res. Pharmacol. Drug Discov. 2, 100026 (2021).
Rahman, M. M. et al. In silico investigation and potential therapeutic approaches of natural products for COVID-19: computer-aided drug design perspective. Front. Cell. Infect. Microbiol. 12, 929430 (2022).
Agrawal, N. & Goyal, A. Potential candidates against COVID-19 targeting RNA-Dependent RNA polymerase: A comprehensive review. Curr. Pharm. Biotechnol. 23, 396–419 (2022) .
Bekheit, M. S., Panda, S. S. & Girgis, A. S. Potential RNA-dependent RNA polymerase (RdRp) inhibitors as prospective drug candidates for SARS-CoV-2. Eur. J. Med. Chem. 252, 115292 (2023).
Eydoux, C. et al. A fluorescence-based high throughput-screening assay for the SARS-CoV RNA synthesis complex. J. Virol. Methods. 288, 114013 (2021).
Bai, X. et al. Identifying small-molecule inhibitors of SARS-CoV-2 RNA-Dependent RNA polymerase by establishing a fluorometric assay. Front. Immunol. 13, 844749 (2022).
Bertolin, A. P. et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp12/7/8 RNA-dependent RNA polymerase. Biochem. J. 478, 2425–2443 (2021).
Zhao, J. et al. Quinoline and Quinazoline derivatives inhibit viral RNA synthesis by SARS-CoV-2 RdRp. ACS Infect. Dis. 7, 1535–1544 (2021).
Li, Q. et al. Corilagin inhibits SARS-CoV-2 replication by targeting viral RNA-dependent RNA polymerase. Acta Pharm. Sin B. 11, 1555–1567 (2021).
Zhang, G. N. et al. Discovery and optimization of 2-((1H-indol-3-yl)thio)-N-benzyl-acetamides as novel SARS-CoV-2 RdRp inhibitors. Eur. J. Med. Chem. 223, 113622 (2021).
Eltahla, A. A., Lackovic, K., Marquis, C., Eden, J. S. & White, P. A. A fluorescence-based high-throughput screen to identify small compound inhibitors of the genotype 3a hepatitis C virus RNA polymerase. J. Biomol. Screen. 18, 1027–1034 (2013).
Sáez-Álvarez, Y., Arias, A., Del Águila, C. & Agudo, R. Development of a fluorescence-based method for the rapid determination of Zika virus polymerase activity and the screening of antiviral drugs. Sci. Rep. 9, 5397 (2019).
Sáez-Álvarez, Y. et al. Novel nonnucleoside inhibitors of Zika Virus polymerase identified through the screening of an Open Library of Antikinetoplastid compounds. Antimicrob. Agents Chemother. 65, e0089421 (2021).
García-Zarandieta, M. et al. Identification of West Nile virus RNA-dependent RNA polymerase non-nucleoside inhibitors by real-time high throughput fluorescence screening. Antiviral Res. 212, 105568 (2023).
Kocabas, F., Turan, R. D. & Aslan, G. S. Fluorometric RdRp assay with self-priming RNA. Virus Genes. 50, 498–504 (2015).
Hs, L. et al. Simeprevir Potently suppresses SARS-CoV-2 replication and synergizes with Remdesivir. ACS Cent. Sci. 7, 792–802 (2021).
Yin, W. et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 28, 319–325 (2021).
Madru, C. et al. Fast and efficient purification of SARS-CoV-2 RNA dependent RNA polymerase complex expressed in Escherichia coli. PloS One. 16, e0250610 (2021).
Tse, W. C. & Boger, D. L. A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Acc. Chem. Res. 37, 61–69 (2004).
del Villar-Guerra, R., Gray, R. D., Trent, J. O. & Chaires, J. B. A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand–nucleic acid structural selectivity. Nucleic Acids Res. 46, e41 (2018).
Wicks, S. L. & Hargrove, A. E. Fluorescent indicator displacement assays to identify and characterize small molecule interactions with RNA. Methods San Diego Calif. 167, 3–14 (2019).
Zhang, J., Umemoto, S. & Nakatani, K. Fluorescent indicator displacement assay for ligand-RNA interactions. J. Am. Chem. Soc. 132, 3660–3661 (2010).
Mravinec, M., Bajc, G. & Butala, M. Surface plasmon resonance approach to study drug interactions with SARS-CoV-2 RNA-dependent RNA polymerase highlights treatment potential of suramin. J. Virol. Methods. 298, 114283 (2021).
Dragan, A. I. et al. SYBR Green I: Fluorescence properties and interaction with DNA. J. Fluoresc. 22, 1189–1199 (2012).
Brant, A. C., Tian, W., Majerciak, V., Yang, W. & Zheng, Z. M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell. Biosci. 11, 136 (2021).
Gul, S. & Gribbon, P. Exemplification of the challenges associated with utilising fluorescence intensity based assays in discovery. Expert Opin. Drug Discov. 5, 681–690 (2010).
Zou, L., Harkey, M. R. & Henderson, G. L. Effects of intrinsic fluorescence and quenching on fluorescence-based screening of natural products. Phytomedicine Int. J. Phytother Phytopharm. 9, 263–267 (2002).
Cruz-León, S. & Schwierz, N. R. N. A. Captures more cations than DNA: Insights from Molecular Dynamics simulations. J. Phys. Chem. B. 126, 8646–8654 (2022).
Pan, F., Roland, C. & Sagui, C. Ion distributions around left- and right-handed DNA and RNA duplexes: A comparative study. Nucleic Acids Res. 42, 13981–13996 (2014).
Wiedemar, N., Hauser, D. A. & Mäser, P. 100 years of Suramin. Antimicrob. Agents Chemother. 64, e01168–e01119 (2020).
Li, X. et al. Corilagin, a promising medicinal herbal agent. Biomed. Pharmacother Biomedecine Pharmacother. 99, 43–50 (2018).
Izquierdo, L. et al. Simeprevir for the treatment of hepatitis C virus infection. Pharmacogenomics Pers. Med. 7, 241–249 (2014).
Vaidya, A. & Perry, C. M. Simeprevir: First global approval. Drugs. 73, 2093–2106 (2013).
Nichols, S. E., Swift, R. V. & Amaro, R. E. Rational prediction with molecular dynamics for hit identification. Curr. Top. Med. Chem. 12, 2002–2012 (2012).
Pirolli, D. et al. Virtual screening and molecular dynamics simulations provide insight into repurposing drugs against SARS-CoV-2 variants spike protein/ACE2 interface. Sci. Rep. 13, 1494 (2023).
Dey, D., Ramakumar, S. & Conn, G. L. Targeted redesign of suramin analogs for novel antimicrobial lead development. J. Chem. Inf. Model. 61, 4442–4454 (2021).
Ono, K., Nakane, H. & Fukushima, M. Differential inhibition of various deoxyribonucleic and ribonucleic acid polymerases by suramin. Eur. J. Biochem. 172, 349–353 (1988).
Jindal, H. K., Anderson, C. W., Davis, R. G. & Vishwanatha, J. K. Suramin affects DNA synthesis in HeLa cells by inhibition of DNA polymerases. Cancer Res. 50, 7754–7757 (1990).
De Clercq, E. Suramin: A potent inhibitor of the reverse transcriptase of RNA tumor viruses. Cancer Lett. 8, 9–22 (1979).
Boniardi, I. et al. Suramin inhibits SARS-CoV-2 nucleocapsid phosphoprotein genome packaging function. Virus Res. 336, 199221 (2023).
Guo, C., Xu, H., Li, X., Yu, J. & Lin, D. Suramin Disturbs the Association of the N-Terminal Domain of SARS-CoV-2 nucleocapsid protein with RNA. Mol. Basel Switz. 28, 2534 (2023).
Sammons, R. M. et al. High-throughput assay for identifying Diverse antagonists of the binding Interaction between the ACE2 receptor and the dynamic spike proteins of SARS-CoV-2. ACS Infect. Dis. 8, 2259–2270 (2022).
Kwon, P. S. et al. Suramin binds and inhibits infection of SARS-CoV-2 through both spike protein-heparan sulfate and ACE2 receptor interactions. Commun. Biol. 6, 387 (2023).
Bibi, N. et al. Drug repositioning against COVID-19: A first line treatment. J. Biomol. Struct. Dyn. 40, 12812–12826 (2022).
J, Z. et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 478, (2021).
Salgado-Benvindo, C. et al. Suramin inhibits SARS-CoV-2 infection in Cell Culture by interfering with early steps of the replication cycle. Antimicrob. Agents Chemother. 64, e00900–e00920 (2020).
Petcu, D. J., Aldrich, C. E., Coates, L., Taylor, J. M. & Mason, W. S. Suramin inhibits in vitro infection by duck hepatitis B virus, Rous sarcoma virus, and Hepatitis delta virus. Virology. 167, 385–392 (1988).
Garson, J. A., Lubach, D., Passas, J., Whitby, K. & Grant, P. R. Suramin blocks hepatitis C binding to human hepatoma cells in vitro. J. Med. Virol. 57, 238–242 (1999).
Albulescu, I. C. et al. Suramin inhibits Chikungunya Virus replication by interacting with virions and blocking the early steps of infection. Viruses. 12, 314 (2020).
Henß, L. et al. Suramin is a potent inhibitor of Chikungunya and Ebola virus cell entry. Virol. J. 13, 149 (2016).
Albulescu, I. C., Kovacikova, K., Tas, A., Snijder, E. J. & van Hemert, M. J. Suramin inhibits Zika virus replication by interfering with virus attachment and release of infectious particles. Antiviral Res. 143, 230–236 (2017).
Tan, C. W., Sam, I. C., Chong, W. L., Lee, V. S. & Chan, Y. F. Polysulfonate suramin inhibits Zika virus infection. Antiviral Res. 143, 186–194 (2017).
Wang, Y., Qing, J., Sun, Y. & Rao, Z. Suramin inhibits EV71 infection. Antiviral Res. 103, 1–6 (2014).
Langendries, L., Abdelnabi, R., Neyts, J. & Delang, L. Repurposing drugs for Mayaro Virus: Identification of EIDD-1931, Favipiravir and Suramin as Mayaro Virus inhibitors. Microorganisms. 9, 734 (2021).
Ogden, A., Wientjes, M. G. & Au, J. L. S. Suramin as a chemosensitizer: Oral pharmacokinetics in rats. Pharm. Res. 21, 2058–2063 (2004).
Mourón, S. et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 (2013).
Dejmek, M. et al. Non-nucleotide RNA-Dependent RNA polymerase inhibitor that blocks SARS-CoV-2 replication. Viruses. 13, 1585 (2021).
Acknowledgements
We are grateful to all members of the DNA Replication and Chromatin Dynamics groups (CNIO) for stimulating discussions. We thank Dr. Marc Delarue and Dr. Ludovic Sauguet for the pRSFDuet-1(nsp8/nsp7)(nsp12) plasmid, Dr. Luis Blanco for the pET16-10His-PrimPol plasmid, Dr. Araceli Grande and Dr. Miguel de Vega kindly providing purified Pol gamma and Φ29pol, respectively. We are also thank Dr. Joaquín Pastor and the CNIO Experimental Therapeutics Programme for discussions and providing sildenafil and duvesilib. Research in JM lab was supported by the Ministry of Science and Innovation, Spain (MCIN/AEI/10.13039/501100011033 grants PID2019-106707-RB and PID2022-142177NB-100; co-sponsored by ERDF A way of making Europe).
Author information
Authors and Affiliations
Contributions
SL: experimental work, study design, manuscript writing. BDG: study design, selection of putative RdRp inhibitors. EC, EB-R, RF-L: experimental contribution to RdRp purification. JM: study design, funding acquisition, manuscript writing. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Llanos, S., Di Geronimo, B., Casajús, E. et al. Interference of small compounds and Mg2+ with dsRNA-binding fluorophores compromises the identification of SARS-CoV-2 RdRp inhibitors. Sci Rep 14, 28250 (2024). https://doi.org/10.1038/s41598-024-78354-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-78354-x