Abstract
The forced expression system of MYOD1, a master gene for myogenic differentiation, can efficiently and rapidly reproduce muscle differentiation of human induced pluripotent stem cells (hiPSCs). Despite these advantages of the MYOD1 overexpression system, developed myotubes are relatively immature and do not recapitulate several aspects of striated muscle fibers. Here, we developed a simple optimized protocol using an alternative culture medium for maximizing the advantages of the MYOD1 overexpression system, and successfully improved the formation of multinucleated mature myotubes within 10 days. In this study, we generated hiPSCs derived from healthy donors and an individual with congenial muscular dystrophy caused by LMNA mutation (laminopathy), and compared disease-associated phenotypes in differentiated myotubes generated by the conventional method and by our new optimized culture method. Using our optimized method, abnormal myonuclear shape was pronounced in the patient-derived iPSCs. In addition, abnormal accumulation of the nuclear membrane protein emerin was observed in LMNA-mutant hiPSCs. Our new culture method is expected to be widely applicable as a MYOD1 overexpression model of hiPSC-derived skeletal muscle cells for the analysis of a variety of muscle diseases.
Similar content being viewed by others
Introduction
Recent advances in induced pluripotent stem cell (iPSC) technology have led to a better understanding of the underlying pathological mechanisms of intractable diseases including muscular dystrophies, and the development of novel methods of regenerative medicine1. Human iPSC (hiPSC) has the ability to differentiate into multiple cell types, and the generation of patient- or disease-specific iPSCs can potentially reproduce the correlation between a genetic mutation and the disease phenotype more precisely than existing cell lines and animal models2. To date, several transgenic and nontransgenic approaches have been developed to generate differentiated skeletal muscle cells from hiPSCs, as well as to enhance the maturity and functionality of muscle cells by adapting the culture conditions3,4,5. The forced expression of master transcription factors for skeletal muscle development (PAX3 or PAX7)6,7, or a master regulator of skeletal muscle lineage (MYOD1)8,9,10, can directly induce hiPSCs to differentiate into skeletal muscle cells. Despite the advantages of such transgenic procedures for the efficient and rapid induction of hiPSCs into muscle cells, promoting the differentiation of hiPSCs into mature myotubes using conventional protocols remains a challenge. Previous studies applied bioengineering culture techniques, such as 3D bioprinting architecture11,12 or electrical-field stimulation13, to successfully improve the degree of myotube maturation; however, these skeletal muscle tissue engineering techniques require a complicated method or an electrical device, and hence inducing myotube maturation in cultured cells remains technically room for improvement4.
In the case of the muscular dystrophies, the generation of patient-derived iPSCs will reflect the actual genetic mutation, and can be used to elucidate genotype-phenotype correlations. These patient-specific cells are a suitable candidate for disease modeling, analysis of myogenic capacity, and elucidation of the underlying pathological mechanism before the onset of typical secondary effects, such as inflammation and fibrosis14. A number of studies have focused on iPSCs derived from patients with Duchenne muscular dystrophy (DMD), using both transgene-based and transgene-free differentiation protocols8,15,16. The modification of the dystrophin gene mutations by CRISPR-Cas9-based genome editing was reported to successfully rescue dystrophin expression in patient-derived iPSCs17, and to improve DMD phenotypes, such as the increase in creatine kinase level18 and muscle damage from contractions caused by electrical stimulation13. Therefore, muscle disease models based on iPSC technology have provided a reliable platform for investigating the pathological mechanisms of a specific mutation.
Mutations in LMNA, encoding lamin A/C, which are proteins localized in the nucleus, result in a variety of diseases collectively called the laminopathies19. More than a dozen types of rare disorders caused by LMNA mutations have been reported to date20,21, and these striated muscle laminopathies include a range of muscular dystrophies, such as Emery–Dreifuss muscular dystrophy (EDMD), limb-girdle muscular dystrophy type 1B (LGMD1B), and congenital muscular dystropohy. Lamin A/C, together with other types of lamin, such as B1 and B2, form flexible fibrous mesh-like structures to provide mechanical support to the nucleus, and play important roles in chromatin organization and signaling pathways22,23. Although the genetic mutations that cause the laminopathies have been identified, the underlying pathological mechanisms of the tissue-restricted disease phenotypes have not yet been elucidated.
Recent studies have reported the generation of skeletal muscle models from laminopathy patient-derived iPSCs11,24. Skeletal muscle differentiation from 3 LMNA-mutant hiPSCs (LMNA L35P, R249W, and K32del) showed abnormalities in myonuclear shape by a standard monolayer culture method, which was more prominent by a 3D artificial muscle culture technique using fibrin hydrogels11. Among the several types of nuclear deformities, the proportion of abnormally elongated nuclei was more pronounced in all 3 hiPSCs of the skeletal muscle laminopathies. Another study from the same group demonstrated that the degree of skeletal muscle differentiation of LMNA-mutant hiPSCs was similar to healthy controls, and disease-associated hallmarks included the presence of nuclear abnormalities and mislocalization of lamin A/C and lamin B124.
In the present study, we established a new monolayer culture method based on the selection of suitable culture media to maximize the advantages of a previously established MYOD1 overexpression strategy10,25. Healthy control hiPSCs, laminopathy patient-derived iPSCs (E33del) and its isogenic control iPSCs generated by CRISPR-Cas9 genome editing were differentiated into myotubes using this optimized method without applying any bioengineering culture techniques. Our optimized method clearly and reproducibly accelerated the formation of multi nucleated mature myotubes derived from the hiPSC lines. In addition, abnormal accumulation of the nuclear membrane protein emerin was confirmed in the LMNA-mutant hiPSCs, recapitulating a characteristic feature of striated muscle laminopathy.
Results
Muscle differentiation was improved by an optimized culture protocol
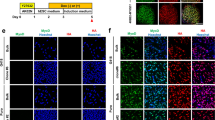
To investigate whether the maturation of skeletal muscle cells could effectively be induced by replacing the standard 2% horse serum (HS)/DMEM differentiation medium to a different medium, several patterns of culture protocols were tested using commercially available myotube medium and myotube fusion medium12. Healthy hiPSCs were plated on Matrigel-coated plates with StemFit on the first day, and switched to PECM medium for an additional 3 days. Doxycycline (Dox) treatment was started on the last day of culture in PECM medium, and continued throughout the following culture protocols. Compared with a culture protocol using 2% HS/DMEM for the next 6 days (the standard protocol) (Fig. 1A), myotube formation was clearly robust when fusion medium (SKM-03+) was used after 2 days of culturing the cells with 2% HS/DMEM (the optimized protocol) (Fig. 1E). This 2-day culture period in 2% HS/DMEM was crucial, as the viability of muscle cells substantially decreased when fusion medium was used immediately after the PECM medium (Fig. 1C). When the myotube medium (SKM-03) was used on day 6 after 2 days of culture in 2% HS/DMEM, the progression of muscle cell differentiation was indistinguishable from the standard protocol (Fig. 1D). The muscle cell culture became unsuccessful on day 10 when the myotube medium was continuously used without the prior 2-day culture in 2% HS/DMEM (Fig. 1B). In addition, when the cells were cultured in the myotube medium for 2 days and then switched to the fusion medium for the following 4 days without using 2% HS/DMEM, the majority of myotubes were gradually lost (Fig. 1F). The optimized culture protocol reproducibly induced more efficient maturation of myotubes from hiPSCs compared with the standard culture protocol (Supplemental Video 1). Morphological changes were observed on the second day of culture in the fusion medium, and myotubes gradually became thicker and more robust. These results indicated that the combination of culture media is crucial for improving the fusion of muscle cells from Dox-inducible MYOD1-hiPSCs.
Schematic diagrams of tested culture protocols using Healthy hiPSCs. (A) The standard protocol using 2%HS/DMEM after day 4 of culture. (B) A protocol using myotube medium on day 4. Cell culture became unsuccessful on day 10. (C) A protocol using fusion medium on day 4. Cell culture became unsuccessful on day 10. (D) A protocol using 2%HS/DMEM and myotube medium. (E) An optimized protocol using 2%HS/DMEM and fusion medium. (F) A protocol using myotube medium and fusion medium. Cell culture was failed at day 10. Y represents Y-27632 (ROCK inhibitor), and Dox represents doxycycline. Scale bar = 500 μm.
Confirmation of myotube maturation using laminopathy patient-derived and CRISPR-Cas9-rescued hiPSCs
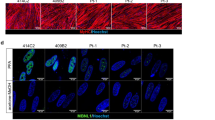
Our optimized culture protocol also effectively induced improved maturation of myotubes in the other two healthy hiPSC lines (Supplemental Fig. 1), the LMNA hiPSCs, and the Rescue hiPSCs (Fig. 2A). The optimized protocol successfully promoted the myotube differentiation which was represented by significantly increase in the myocyte induction efficiency, calculated by the percentage of nuclei in the MHC-positive area (Fig. 2B). The degree of myotube maturation was assayed by calculating the expression levels of muscle-related proteins and the number of nuclei in the myotubes. The protein levels of dystrophin, MHC, and actinin were increased in the optimized protocol in the three cell lines of Healthy, LMNA, and Rescue hiPSC by Western blotting (Fig. 2C,D). The majority of myotubes had only one or two nuclei in all three cell lines cultured with the standard protocol, whereas the percentage of myotubes with a single nucleus was significantly decreased in the respective hiPSC lines on day 10 of culture using the optimized protocol (Fig. 2E). The optimized protocol improved multi nucleation, and hence Healthy hiPSCs cultured using this protocol had significantly higher frequencies of myotubes with 3, 4, 5, and 6 nuclei compared with Healthy hiPSCs cultured using the standard protocol, and similar trends were observed in the LMNA and the Rescue hiPSCs (Fig. 2E). In addition, the efficacy of skeletal muscle cell differentiation was evaluated by the expression of maturation-associated markers using quantitative PCR on day 10 of muscle differentiation. Although the embryonic MHC gene MYH3 expression was still significantly increased in both the LMNA and the Rescue hiPSCs (Fig. 2F), other maturation-associated markers, such as MYH7 (the slow MHC gene), MYH2 (the fast-oxidative MHC gene), ATP2A1 and CASQ1 (genes associated with the development of the sarcoplasmic reticulum), and TMEM8C (muscle-specific membrane activator of myoblast fusion) were significantly increased in all three cell lines using the optimized protocol (Fig. 2F). The degree of upregulation of differentiation-associated genes was variable among the three cell lines, but these findings indicated highly improved efficacy of muscle maturation of the MYOD1 overexpression model of hiPSCs using the optimized protocol.
Promotion of muscle maturation of Healthy, LMNA-mutant, and LMNA-rescue hiPSCs using the optimized protocol. (A) Representative immunohistochemical staining for MHC in the differentiated hiPSC lines cultured for 10 days with the standard or optimized protocol. Scale bar = 100 μm. (B) The myocyte induction efficacy calculated by the percentage of nuclei in the MHC-positive area. Five images (×20 objectives) were taken from each well of the three cell lines and analyzed. (C) Images of Western blots performed on three individual samples to quantify the levels of dystrophin, MHC, and actinin in the three cell lines. (D) Graph showing the relative expression of each protein to the levels of lamin B1 expression. (E) Frequency of myotubes with specific number of nuclei as an indicator of muscle maturation. Four to five images (×10 objectives) were taken from each well of the 3 cell lines and analyzed. (F) Quantification of genes associated with muscle differentiation by RT-qPCR. Relative mRNA levels of MYH3, MYH7, MYH2, ATP2A1, CASQ1, and TMEM8C were normalized to the level of TBP, and shown as the fold increase to cells cultured using the standard protocol. Data are shown as the mean ± SD (n = 3, each group). *P < 0.05, **P < 0.01, and ***P < 0.001 versus the standard protocol.
Enhancement of myonuclear abnormalities of laminopathy in hiPSCs using the optimized culture protocol
Repeatedly, both LMNA and Rescue hiPSCs morphologically resembled Healthy hiPSCs regarding proliferation, and all three cell lines successfully differentiated with similar dynamics to form multinucleated myotubes using the optimized protocol; therefore, we applied our optimized protocol to study the pathological features of laminopathy. The presence of abnormally shaped nuclei in skeletal muscle cells is a hallmark of the laminopathies20,26. Differentiated myotubes were immunostained for MHC and lamin A/C (Fig. 3A) to quantify the percentage of nuclear shape deformities and the length of the myonuclei. In LMNA-mutant myotubes, mildly deformed nuclei, i.e., nuclei with blebbing, nuclei with a string part, and nuclei that were abnormally elongated were typically observed (Fig. 3B). The percentage of these abnormal nuclei was 11.7% ± 1.8% in LMNA hiPSCs using the standard protocol, which was significantly increased using the optimized protocol (18.2% ± 2.8%, P < 0.001 vs. LMNA iPSCs using the standard protocol). Few abnormal nuclei were detected in differentiated muscle cells from Healthy and Rescue hiPSCs, and the percentages of abnormal nuclei were unchanged between cells grown in the standard protocol and those grown in the optimized protocol (Fig. 3C). As severely elongated nuclei and nuclei with a string part were not observed in myotubes from Healthy and Rescue hiPSCs, these abnormalities appear to be specific features of LMNA hiPSCs. Furthermore, the average length of the major axis of myonuclei was significantly elongated in LMNA hiPSCs compared with Healthy and Rescue hiPSCs cultured with either the standard protocol or the optimized protocol (Fig. 3D), whereas the optimized protocol contributed to the significant increase in the percentage of elongated myonuclei in LMNA hiPSCs (P < 0.001 vs. the standard protocol). In addition to abnormalities in nuclear shape, abnormalities in the localization of nuclear envelope (NE) proteins were observed in the cells cultured with the optimized protocol, as observed by immunofluorescence staining. The localizations of lamin A/C, emerin, NUP153, and LAP2 were maintained in the bleb, string, and elongated regions of myonuclei, whereas the expression of lamin B1 was absent in the myonuclear blebs of LMNA hiPSCs (Fig. 4A). Interestingly, the inner NE protein emerin was highly accumulated in the blebs of myonuclei from LMNA-mutant myotubes, and this was also observed in the nuclear blebs of cells cultured using the standard protocol. Moreover, Matrigel-coated glass-bottom dishes and silicone chambers were used to compare the effects of stiffness of the culture base materials on emerin expression in the blebs (Fig. 4B). The accumulation of emerin was similarly observed in the blebs from LMNA hiPSCs cultured on either a hard glass surface or a soft silicone surface (Supplemental Fig. 2). These results indicate that the mislocalization of NE proteins in the abnormally deformed parts of myonuclei depend on the type of NE protein. The accumulation of emerin in the blebs was constantly observed in LMNA hiPSCs, independent of culture protocols and culture platforms.
Increased nuclear abnormalities in LMNA-mutant hiPSCs using the optimized protocol. (A) Immunohistochemical staining for MHC and lamin A, with a DAPI nuclear counterstain, for the detection of abnormalities in nuclear shape in Healthy, LMNA, and Rescue hiPSCs using the standard and optimized protocols. Scale bar = 100 μm. (B) Representative images of the abnormal shapes of nuclei stained with lamin A, characterized as blebs, mild deformation, elongation, stringing, and severe stringing using the optimized protocol. Each panel displays individual black and white images. Yellow arrows indicate representative nuclear abnormalities. Scale bar = 40 μm. (C) Graph showing percentages of myonuclei with abnormal shapes, and (D) graph showing average lengths of the major axis of nuclei within myotubes, calculated to evaluate myonuclear lengthening in the 3 hiPSC lines, cultured using either the standard or the optimized protocol. Data were obtained from 300 to 450 myonuclei of each cell line. Data are shown as the mean ± SD. ***P < 0.001.
Localization of NE proteins in nuclei from differentiated myotubes of LMNA-mutant iPSCs. (A) Lamin B1, emerin, NUP153, and LAP2 were double-stained with lamin A/C and a DAPI nuclear counterstain. Yellow arrows indicate the representative localizations of nuclear envelope proteins in abnormal regions of myonuclei. (B) Accumulation of emerin expression in abnormal regions of myonuclei from LMNA hiPSCs cultured on a glass-bottom dish, a plastic-bottom dish, and a silicone chamber. Scale bar = 50 μm.
Discussion
In this study, we tested several culture protocols to find an improved alternative protocol to the basic muscle differentiation method using MYOD1-overexpressed hiPSCs. In our improved method, the application of a specific combination of commercially available media at specific timings is critical. Our newly developed optimized protocol successfully improved skeletal muscle maturation with high reproducibility, which was confirmed by testing several hiPSC lines, and hence maximized the benefits of the MYOD1-mediated muscle differentiation approach. Myotubes cultured using the optimized protocol were multi nucleated and showed increased levels of muscle related proteins and gene expression of various MYH subtypes within 10 days of culture.
Since the discovery of iPSCs in 20061, remarkable progress has been reported in the generation of myogenic cells from hiPSCs. The culture conditions were modified and adapted using growth factors and various kinds of media in each report. In the field of skeletal muscle development using hiPSCs, transgenic and nontransgenic approaches for generating differentiated muscle cells from hiPSCs have been shown to have both advantages and disadvantages27,28. The forced expression of transcription factors can effectively generate muscle cells with high purity, although the myotubes are immature and show neither multi nucleation nor sarcomeric structures. In addition, myotube maturation can be promoted using bioengineering culture techniques, such as 3D bioprinting architecture and electric-field stimulation, although specific devices are necessary to use these techniques in the experimental setting4. Compared with methods using transgenic approaches, direct muscle differentiation protocols require longer times as the cells need to undergo several differentiation stages29; however, it is possible to generate mature myotubes, which are able to undergo spontaneous self-contractions5.
The clinical severity of LMNA-associated muscular dystrophies are evaluated by the levels of skeletal muscle weakness and atrophy, cardiac dysfunction, joint contractures, and spinal deformities30,31. Regarding the laminopathies, understanding their tissue-specific phenotypes is particularly important toward gaining a better understanding of the specific roles of lamin A/C in different tissues, although the genetic and pathogenic mechanisms of these diseases still remain unclear. Owing to the rarity of the skeletal muscle laminopathies, only a small amount of data from studies using patient-derived iPSCs is available. Recent studies established and analyzed 3 MYOD1-overexpressed LMNA-mutant hiPSCs (LMNA L35P, R249W, and K32del) by applying a 3D artificial skeletal muscle platform using fibrin hydrogels11,24. This tissue engineering technique enabled the recapitulation of the nuclear shape abnormalities in differentiated myotubes, in which nuclear elongation was the predominant abnormality, consistent with previous reports using muscle biopsies and primary myoblasts from laminopathy patients32,33. However, it is still challenging to apply these bioengineering culture techniques for high-throughput drug screening25.
Here, we established a novel laminopathy patient-derived iPSC line (E33del) and its isogenic control generated by CRISPR-Cas9 technology, using their direct differentiation into myogenic cells by MYOD1 overexpression. We clearly demonstrated that nuclear shape abnormalities, including nuclear elongation, were specific phenotypes in the LMNA hiPSCs, which were recovered in the Rescue hiPSCs cultured by the standard protocol. Moreover, promoting myotube maturation using the optimized protocol increased the percentage of nuclear abnormalities by up to 18% in the patient-derived iPSCs. In skeletal muscle sections from EDMD/LGMD1B patients, about 20% of myonuclei have irregular shapes34; therefore, applying the optimized protocol can recapitulate the abnormalities of skeletal muscle laminopathies, even at the monolayer culture level. These unique nuclear phenotypes are thought to result from the increased fragility, altered stiffness, and irregular localization of highly condensed heterochromatin, as demonstrated by studies using murine models of the laminopathies35,36,37; however, the underlying mechanisms and correlation with clinical phenotypes still remain unclear. One benefit of using LMNA patient-derived iPSCs to study the genotype-phenotype correlations is that nuclear shape abnormalities can be analyzed in differentiated muscle cells. In contrast to the impaired muscle cell differentiation that is observed in primary muscle stem cells from laminopathy patients (K32del, L380S, and R249W)38, myotube fusion and differentiation were well-maintained in the LMNA hiPSCs, which were similarly reported in previous studies11,24. We also detected the mislocalization of NE proteins in nuclei with morphological defects from differentiated LMNA-mutant myotubes. In our model, lamin A/C was localized evenly in myotube nuclei with normal, elongated, blebs, and string shapes, whereas lamin B1 was not observed within nuclear blebs. The uneven distribution of lamin A/C and lamin B1 was also reported in iPS-mesenchymal stem cells derived from patients with Hutchinson-Gilford progeria syndrome, which is caused by point mutations in the LMNA gene39, and in fibroblasts from patients with familial partial lipodystrophy of the Dunnigan type, which is caused by missense heterozygous mutations in the LMNA gene40. Other NE proteins, such as NUP153 located in the nuclear pore complex, and LAP2 located in the inner nuclear membrane were localized throughout the entire nuclei, including in the abnormal nuclear regions. The localization of emerin was the most prominent feature of NE proteins expressed in abnormal myonuclei from laminopathy patient-derived iPSCs. Emerin is a ubiquitously expressed nuclear inner membrane protein41, and mutations in the emerin gene cause EDMD42. Emerin directly interacts with lamin A/C43,44, and the depletion of these NE proteins affects chromatin organization in cancer cells in vitro45. Moreover, recent studies clearly demonstrated that emerin plays key roles in front-rear cell polarity in myoblasts46, and in nuclear migration to reseal injured muscle membranes47. In LMNA hiPSCs, the accumulation of emerin was particularly detected in the protruded regions, and on 1 side of differentiated myonuclei, which is similarly observed in cells seeded onto plastic-bottom, glass-bottom, and silicone-bottom culture materials. Although it is still unknown whether the accumulation of emerin contributes to the progression of nuclear abnormalities or to protection of the nuclear membrane against mechanical stress, the reproduction of abnormal myonuclear shapes in differentiated myotubes using LMNA hiPSCs could provide the foundations for elucidating the pathological mechanisms of laminopathy. Moreover, our simple optimized protocol for skeletal myogenic differentiation of hiPSCs can be applied for high-throughput screening to investigate potential therapeutic drugs in the future.
In conclusion, we established an optimized monolayer myogenic differentiation culture method to maximize the advantages of a previously established MYOD1 overexpression strategy. This new approach was effective for developing more mature myotubes using 5 different hiPSC lines, including a new skeletal muscle laminopathy model and its isogenic control; therefore, our approach can be applied to other MYOD1-overexpressed hiPSCs derived from patients with various muscle diseases. The limitations of this study include the utilization of the Dox-inducible MYOD1-overexpression system, and the existence of unknown components in the myotube fusion medium used in the optimized protocol. It is unclear whether the protocol can improve the muscle differentiation of hiPSCs generated by other transgenic approaches or direct myogenic induction systems without transgenes. Taken together, our monolayer culture approach is beneficial for improving myotube formation, and is easy to adapt for studying other muscular dystrophy-hiPSCs models.
Materials and methods
Generation and maintenance of hiPSC lines
The hiPSC line 414C248, 409B2 line48, MYOD1‑hiPSC25, a LMNA-mutant hiPSC line and its isogenic control line (Rescue) were used. The 414C2 line was used as the Healthy control hiPSCs for comparing with the LMNA-mutant and LMNA-rescue iPSCs. The hiPSC line 414C2, 409B2, and MYOD1‑hiPSC were generated from human fibroblast cells purchased from Cell Applications, Inc., (USA). The LMNA-mutant (LMNA) hiPSC line was generated using episomal vectors transfected into dermal fibroblasts from a LMNA-related congenital muscular dystrophy patient with heterozygous mutations (c.99-101delGGA, p.E33del) (Supplemental Fig. 3A). To generate the Rescue line, CRISPR-Cas9 genome editing and homology template-dependent repair were performed to correct the “GGA” trinucleotide deletion at exon 1 of the LMNA gene. Briefly, the Cas9 protein (5 µg), a gRNA (1.5 µg) targeting the mutant LMNA gene (target sequence: GCTGCAGGAGAAGGA***CCTGCaGG, where “***” represents the deleted trinucleotide), and a donor template DNA vector (0.5 µg) with a normal LMNA sequence and a floxed puromycin resistance cassette were electroporated into the LMNA-mutant hiPSCs using 4D Nucleofector. After puromycin (1 µg/mL) selection, successfully targeted subclones were isolated49. Finally, the puromycin resistance cassette was removed by the transient expression of Cre recombinase50 (Supplemental Fig. 3B). To induce the myogenic differentiation of hiPSCs, these cells were used to generate Dox-inducible MYOD1-hiPSCs at Center for iPS Cell Research and Application (CiRA) at Kyoto University, as described previously10,51. The cells were cultured on a feeder-free system using laminin-coated (iMatrix-511; Nippi, Japan) 6-well plates (IWAKI, Japan) at the density of 6.5 × 104 cells/well in StemFit AK02N culture media (Reprocell, Japan) containing 10 µM Y-27632 (StemFit + Y) for proliferation.
Skeletal muscle differentiation from hiPSCs
Cells were passaged once a week using Accutase cell dissociation reagent (Nakarai, Japan), and were counted and seeded onto growth factor reduced Matrigel (Corning, USA)-coated 6-well plates in StemFit + Y-27632 at a density of 1 × 104 cells/well for the Healthy hiPSCs and 0.5 × 104 cells/well for the LMNA and the Rescue hiPSCs on the first day after plating. The muscle differentiation protocol of hiPSCs was modified from the previously reported original method10,25. Cells were continuously cultured for another 3 days in Primate ES Cell Medium (PECM) (Reprocell) containing 10 µM Y-27632 (PECM + Y). The myogenic differentiation of hiPSCs was induced by adding 0.3 µg/mL Dox to PECM + Y on day 4, followed by replacing the medium to 2% HS/DMEM supplemented with 0.3 µg/mL Dox, 1% penicillin-streptomycin, 10 ng/mL IGF-1, and 5 µM SB431542. The media were replaced every other day until day 10.
To induce more efficient muscle differentiation, 2% HS/DMEM was changed to myotube medium (SKM-03) or myotube fusion medium (SKM-03+) (Myocea, USA) on day 6 of culture. Schemes of the myogenic differentiation protocols of hiPSCs are shown in Fig. 1.
Bright-field images of differentiated hiPSCs were captured using an ECLIPSE Ts2R microscope (Nikon, Japan). For time-lapse images, Healthy hiPSCs were differentiated into myotubes for 8 days with either 2% HS/DMEM or fusion medium. Sets of time-lapse bright-field images were automatically taken in 30 min intervals for 24 h by IN Cell Analyzer 2200 (Cytiva, USA) equipped with a control system to maintain the cells at 37 °C and 5% CO2.
Immunocytochemistry
Cells were cultured on a Matrigel-coated plastic dish, 35-mm glass-base dish (#3911-035, IWAKI), or silicone chamber (SC4Dea, Menicon, Japan), were fixed with 4% PFA/PBS for 10 min at room temperature. After washing with PBS, cells were permeabilized with 0.5% triton X-100/PBS for 15 min before the immunofluorescence staining protocol was applied. The samples were incubated with primary antibodies at 37 °C for 1 h, and Alexa Fluor 488- or 568-conjugated secondary antibodies (1:500, Thermo Fisher Scientific, USA) with DAPI solution to stain nuclei. The myocyte induction efficiency was determined by calculating the percentage of DAPI-positive nuclei in the myosin heavy chain (MHC)-positive area. Five images (× 20 objectives) were taken from each well and analyzed. Primary antibodies used in this experiment were follows: mouse anti- MHC (MF20; 1:100, DSHB, USA), mouse anti-lamin A/C (4C11; 1:500, Cell Signaling, USA), rabbit anti-lamin A (M52-1; 1:500)52, rabbit anti-emerin (ab156871; 1:300, Abcam, USA), mouse anti-lamin B1 (66095-1-Ig; 1:300, Proteintech, USA), mouse anti-nuclear pore complex 153 (NUP153; ab24700; 1:300, Abcam), and rabbit anti-lamin associated protein 2 (LAP2; ab5162; 1:500, Abcam). Images were visualized and photographed using an Axio Scope A1 microscope (Zeiss, Germany).
Calculation of the fusion index and analysis of nuclear abnormalities
Skeletal muscle cells derived from both protocol using the 2% HS/DMEM and that using myotube fusion medium were analyzed on day 10, for calculating the fusion index, percentage of abnormal nuclei, and myonuclear length. The fusion index (the degree of muscle cell maturation) was calculated as the percentage of nuclei within differentiated myotubes. Nuclear abnormalities were evaluated by the percentage of nuclei with abnormal shapes and lengths. The following abnormal myonuclear morphologies were considered an ‘abnormal shape’: nuclei with blebs (protrusions), nuclei with strings (1 nucleus divided into 2 and linked with a thin bridge), and severely elongated nuclei (major axis > 40 μm), by a blinded researcher, as previously reported24. The elongation of myonuclei was notably observed in LMNA hiPSCs, so the lengths of the major myonuclear axies were calculated using NIH ImageJ software. Imaging fields were randomly selected, and 300–450 myonuclei were analyzed from each cell line. Localizations of the NE proteins lamin A/C, lamin B1, emerin, NUP153, and LAP2, were detected by immunofluorescence staining in cells cultured using the optimized protocol.
RNA isolation and quantitative reverse-transcription PCR (RT-qPCR)
Total RNA was extracted from cultured hiPSCs at 10 days of culture using RNeasy Plus Universal Mini kit (QIAGEN, Germany). The purity and integrity of the extracted RNA were determined using a NanoDrop One spectrophotometer (Thermo Fisher Scientific) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Real-time RT-qPCR was performed using the QuantStudio 3 Real-Time PCR system using SYBR green master mix (Applied Biosystems, USA). The primers used in this study are listed in Table 1. All results were normalized using the TATA-binding protein gene (TBP), and gene expression levels were calculated by the ΔΔCT method of relative quantification. Data were expressed as the fold-increase versus the values of cells cultured using the standard protocol.
Western blot analysis
The cultured hiPSCs were lysed in RIPA buffer containing protease inhibitors and phosphatase inhibitors (Roche). Samples were centrifuged at 15,000 rpm for 15 min, and supernatants were mixed with equal volume of sample buffer solution (Fujifilm).Total proteins were separated on 5–20% and 10–20% gradient SDS-PAGE gels (Fujifilm) and transferred onto PVDF membranes using a semi-dry transfer system (Bio-Rad). After blocking with 2% casein in TBS with 0.05% tween-20 (TBST), primary antibodies were applied to the membranes overnight at 4 °C. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody. All bands were visualized using an ECL substrate solution with ChemiDoc imager (Bio-Rad). The band intensities of the target proteins were analyzed using NIH ImageJ software and were normalized by the band intensity of lamin B1. The primary antibodies used in western blotting were as follows: anti-dystrohin (ab15277, Abcam), anti-skeletal muscle myosin (sc-32732, Santa Cruz, USA), anti-α-actinin (05-387, Merck, Germany), and anti-lamin B1 (66095-1-Ig, Proteintech).
Statistical analysis
Data are shown as the mean ± standard deviation (SD), and differences were determined by 1-way ANOVA with the post-hoc Tukey multiple comparison test or the post-hoc Dunnett t-test. In cases where the means of two parameters were compared, differences were determined by the Welch’s t-test. Statistical tests were 2-sided, and a p-value of less than 0.05 was considered to indicate a statistically significant difference between groups. All statistical analyses were performed using SPSS statistics 28 software (IBM).
Data availability
The datasets analyzed in this study are available from the corresponding author upon reasonable request.
References
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. https://doi.org/10.1016/j.cell.2006.07.024 (2006).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. https://doi.org/10.1016/j.cell.2007.11.019 (2007).
Kodaka, Y., Rabu, G. & Asakura, A. Skeletal muscle cell induction from pluripotent stem cells. Stem Cells Int. 2017 (1376151). https://doi.org/10.1155/2017/1376151 (2017).
Iberite, F., Gruppioni, E. & Ricotti, L. Skeletal muscle differentiation of human iPSCs meets bioengineering strategies: perspectives and challenges. NPJ Regen. Med. https://doi.org/10.1038/s41536-022-00216-9 (2022).
Fujiwara, K. et al. Mature myotubes generated from Human-Induced pluripotent stem cells without forced gene expression. Front. Cell. Dev. Biol. 10, 886879. https://doi.org/10.3389/fcell.2022.886879 (2022).
Darabi, R. et al. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell. Stem Cell. 10, 610–619. https://doi.org/10.1016/j.stem.2012.02.015 (2012).
Quattrocelli, M. et al. Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle. J. Clin. Investig. 125, 4463–4482. https://doi.org/10.1172/JCI82735 (2015).
Goudenege, S. et al. Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol. Ther. 20, 2153–2167. https://doi.org/10.1038/mt.2012.188 (2012).
Maffioletti, S. M. et al. Efficient derivation and inducible differentiation of expandable skeletal myogenic cells from human ES and patient-specific iPS cells. Nat. Protoc. 10, 1457. https://doi.org/10.1038/nprot0915-1457d (2015). Corrigendum.
Tanaka, A. et al. Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi Myopathy in vitro. PLoS One. 8, e61540. https://doi.org/10.1371/journal.pone.0061540 (2013).
Maffioletti, S. M. et al. Three-Dimensional Human iPSC-Derived Artificial skeletal muscles model muscular dystrophies and enable multilineage tissue Engineering. Cell. Rep. 23, 899–908. https://doi.org/10.1016/j.celrep.2018.03.091 (2018).
Pinton, L. et al. 3D human induced pluripotent stem cell-derived bioengineered skeletal muscles for tissue, disease and therapy modeling. Nat. Protoc. 18, 1337–1376. https://doi.org/10.1038/s41596-022-00790-8 (2023).
Uchimura, T., Asano, T., Nakata, T., Hotta, A. & Sakurai, H. A muscle fatigue-like contractile decline was recapitulated using skeletal myotubes from Duchenne muscular dystrophy patient-derived iPSCs. Cell. Rep. Med. 2, 100298. https://doi.org/10.1016/j.xcrm.2021.100298 (2021).
Ortiz-Vitali, J. L. & Darabi, R. iPSCs as a platform for Disease modeling, Drug Screening, and personalized therapy in muscular dystrophies. Cells https://doi.org/10.3390/cells8010020 (2019).
Moretti, A. et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 26, 207–214. https://doi.org/10.1038/s41591-019-0738-2 (2020).
Al Tanoury, Z. et al. Differentiation of the human PAX7-positive myogenic precursors/satellite cell lineage. Development https://doi.org/10.1242/dev.187344 (2020).
Gee, P., Xu, H. & Hotta, A. Cellular Reprogramming, Genome Editing, and Alternative CRISPR Cas9 technologies for Precise Gene Therapy of Duchenne muscular dystrophy. Stem Cells Int. 2017, 8765154. https://doi.org/10.1155/2017/8765154 (2017).
Young, C. S. et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-Derived muscle cells. Cell. Stem Cell. 18, 533–540. https://doi.org/10.1016/j.stem.2016.01.021 (2016).
Wilson, K. L. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell. Biol. 10, 125–129. https://doi.org/10.1016/s0962-8924(99)01708-0 (2000).
Worman, H. J. Nuclear lamins and laminopathies. J. Pathol. 226, 316–325. https://doi.org/10.1002/path.2999 (2012).
Ho, R. & Hegele, R. A. Complex effects of laminopathy mutations on nuclear structure and function. Clin. Genet. 95, 199–209. https://doi.org/10.1111/cge.13455 (2019).
de Leeuw, R., Gruenbaum, Y. & Medalia, O. Nuclear lamins: thin filaments with major functions. Trends Cell. Biol. 28, 34–45. https://doi.org/10.1016/j.tcb.2017.08.004 (2018).
Malashicheva, A. & Perepelina, K. Diversity of Nuclear Lamin A/C action as a key to tissue-specific regulation of Cellular Identity in Health and Disease. Front. Cell. Dev. Biol. 9, 761469. https://doi.org/10.3389/fcell.2021.761469 (2021).
Steele-Stallard, H. B. et al. Modeling skeletal muscle laminopathies using Human Induced Pluripotent stem cells carrying pathogenic. Front. Physiol. 9, 1332. https://doi.org/10.3389/fphys.2018.01332 (2018).
Uchimura, T., Otomo, J., Sato, M. & Sakurai, H. A human iPS cell myogenic differentiation system permitting high-throughput drug screening. Stem Cell. Res. 25, 98–106. https://doi.org/10.1016/j.scr.2017.10.023 (2017).
Stiekema, M., van Zandvoort, M. A. M. J., Ramaekers, F. C. S. & Broers, J. L. V. Structural and mechanical aberrations of the Nuclear Lamina in Disease. Cells https://doi.org/10.3390/cells9081884 (2020).
Jiwlawat, N., Lynch, E., Jeffrey, J., Van Dyke, J. M. & Suzuki, M. Current Progress and Challenges for Skeletal Muscle Differentiation from Human Pluripotent Stem Cells Using Transgene-Free Approaches. Stem Cells Int https://doi.org/10.1155/2018/6241681 (2018).
Yan, L., Rodríguez-delaRosa, A. & Pourquié, O. Human muscle production in vitro from pluripotent stem cells: Basic and clinical applications. Semin Cell. Dev. Biol. 119, 39–48. https://doi.org/10.1016/j.semcdb.2021.04.017 (2021).
Zhao, M. et al. Induced fetal human muscle stem cells with high therapeutic potential in a mouse muscular dystrophy model. Stem Cell. Rep. 15, 80–94. https://doi.org/10.1016/j.stemcr.2020.06.004 (2020).
Maggi, L., Carboni, N. & Bernasconi, P. Skeletal muscle laminopathies: a review of clinical and molecular features. Cells https://doi.org/10.3390/cells5030033 (2016).
Hayashi, Y. K. Nuclear envelope myopathy. Neurol. Clin. Neurosci. 10, 298–303. https://doi.org/10.1111/ncn3.12602 (2022).
Bertrand, A. T. et al. Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J. Cell. Sci. 127, 2873–2884. https://doi.org/10.1242/jcs.144907 (2014).
Tan, D. et al. Phenotype-Genotype Analysis of Chinese Patients with early-onset LMNA-Related muscular dystrophy. PLoS One. 10, e0129699. https://doi.org/10.1371/journal.pone.0129699 (2015).
Park, Y. E. et al. Nuclear changes in skeletal muscle extend to satellite cells in autosomal dominant Emery-Dreifuss muscular dystrophy/limb-girdle muscular dystrophy 1B. Neuromuscul. Disord. 19, 29–36. https://doi.org/10.1016/j.nmd.2008.09.018 (2009).
Earle, A. J. et al. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 19, 464–473. https://doi.org/10.1038/s41563-019-0563-5 (2020).
Wada, E. et al. Deficiency of emerin contributes differently to the pathogenesis of skeletal and cardiac muscles in LmnaH222P/H222P mutant mice. PLoS One. 14, e0221512. https://doi.org/10.1371/journal.pone.0221512 (2019).
Wada, E., Susumu, N., Kaya, M. & Hayashi, Y. K. Characteristics of nuclear architectural abnormalities of myotubes differentiated from Lmna. Vitro Cell. Dev. Biol. Anim. https://doi.org/10.1007/s11626-024-00915-1 (2024).
Owens, D. J. et al. Lamin-related congenital muscular dystrophy alters mechanical signaling and skeletal muscle growth. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22010306 (2020).
Cho, S. et al. Progerin phosphorylation in interphase is lower and less mechanosensitive than lamin-A,C in iPS-derived mesenchymal stem cells. Nucleus 9, 230–245. https://doi.org/10.1080/19491034.2018.1460185 (2018).
Vigouroux, C. et al. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell. Sci. 114, 4459–4468. https://doi.org/10.1242/jcs.114.24.4459 (2001).
Nagano, A. et al. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat. Genet. 12, 254–259. https://doi.org/10.1038/ng0396-254 (1996).
Bione, S. et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8, 323–327. https://doi.org/10.1038/ng1294-323 (1994).
Sakaki, M. et al. Interaction between emerin and nuclear lamins. J. Biochem. 129, 321–327 (2001).
Clements, L., Manilal, S., Love, D. R. & Morris, G. E. Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun. 267, 709–714. https://doi.org/10.1006/bbrc.1999.2023 (2000).
Ranade, D., Pradhan, R., Jayakrishnan, M., Hegde, S. & Sengupta, K. Lamin A/C and emerin depletion impacts chromatin organization and dynamics in the interphase nucleus. BMC Mol. Cell. Biol. 20, 11. https://doi.org/10.1186/s12860-019-0192-5 (2019).
Nastały, P. et al. Role of the nuclear membrane protein emerin in front-rear polarity of the nucleus. Nat. Commun. 11, 2122. https://doi.org/10.1038/s41467-020-15910-9 (2020).
Roman, W. et al. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science 374, 355–359. https://doi.org/10.1126/science.abe5620 (2021).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 8, 409–412. https://doi.org/10.1038/nmeth.1591 (2011).
Xu, H., Kita, Y., Bang, U., Gee, P. & Hotta, A. Optimized electroporation of CRISPR-Cas9/gRNA ribonucleoprotein complex for selection-free homologous recombination in human pluripotent stem cells. STAR. Protoc. 2, 100965. https://doi.org/10.1016/j.xpro.2021.100965 (2021).
Kagita, A. et al. Efficient ssODN-Mediated targeting by avoiding Cellular Inhibitory RNAs through Precomplexed CRISPR-Cas9/sgRNA Ribonucleoprotein. Stem Cell. Rep. 16, 985–996. https://doi.org/10.1016/j.stemcr.2021.02.013 (2021).
Shoji, E. et al. Early pathogenesis of Duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Sci. Rep. 5, 12831. https://doi.org/10.1038/srep12831 (2015).
Navarro, C. L. et al. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum. Mol. Genet. 13, 2493–2503. https://doi.org/10.1093/hmg/ddh265 (2004).
Acknowledgements
We thank Dr. Helena Popiel (Tokyo Medical University) for editing the manuscript and National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP) (Japan) for providing the lamin A antibody (M52-1).
Author information
Authors and Affiliations
Contributions
All authors contributed to design the study and approved the final version of the manuscript. EW: Conceptualization, Formal experiment and analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. NS: Formal experiment and analysis, Investigation, Methodology, Writing–review and editing. YO: Generation of hiPSCs, Investigation, Writing–review and editing. AH: Generation of hiPSCs, Investigation, Writing–review and editing. HS: Generation of hiPSCs, Resources, Investigation, Writing–review and editing. YH: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing–review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary video 1-1
Supplementary video 1-2
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wada, E., Susumu, N., Okuzaki, Y. et al. Optimized simple culture protocol for inducing mature myotubes from MYOD1-overexpressed human iPS cells. Sci Rep 14, 28783 (2024). https://doi.org/10.1038/s41598-024-79745-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-79745-w