Abstract
Soil water deficit (WD) significantly impacts plant survival and crop yields. Many gaps remain in our understanding of the synergistic coordination between molecular and ecophysiological responses delaying substantial drought-induced effects on plant growth. To investigate this synergism in tomato leaves, we combined molecular, ecophysiological, and anatomical methods to examine gene expression patterns and physio-anatomical characteristics during a progressing WD experiment. Four sampling points were selected for transcriptomic analysis based on the key ecophysiological responses of the tomato leaves: 4 and 5 days after WD (d-WD), corresponding to 10% and 90% decrease in leaf stomatal conductance; 8 d-WD, the leaf wilting point; and 10 d-WD, when air embolism blocks 12% of the leaf xylem water transport. At 4 d-WD, upregulated genes were mostly linked to ABA-independent responses, with larger-scale ABA-dependent responses occurring at 5 d-WD. At 8 d-WD, we observed an upregulation of heat shock transcription factors, and two days later (10 d-WD), we found a strong upregulation of oxidative stress transcription factors. Finally, we found that young leaves present a stronger dehydration tolerance than mature leaves at the same drought intensity level, presumably because young leaves upregulate genes related to increased callose deposition resulting in limiting water loss to the phloem, and related to increased cell rigidity by modifying cell wall structures. This combined dataset will serve as a framework for future studies that aim to obtain a more holistic WD plant response at the molecular, ecophysiological and anatomical level.
Similar content being viewed by others
Introduction
The environmental conditions, encompassing all abiotic and biotic factors, define the niche boundaries where a plant species can actively adapt and grow1,2. Any deviation in these conditions from the optimal niche range leads to plant stress and reduced growth and yield3,4. In the present age of anthropogenic climate change, it is evident that global hydrological patterns are swiftly evolving, leading to diminished precipitation in numerous geographical areas that makes them less suitable for agriculture5,6,7,8,9. In a future world where plant growth will become more demanding, there is an urgent need to focus more on mechanisms underlying drought responses in crops to safeguard food production10,11. Tomato (Solanum lycopersicum L.) is one of the major crop species cultivated in the Mediterranean area where droughts are projected to become more intense and more frequent12,13,14. Previous research investigating tomato response to water deficit (WD) is scattered and does not provide a complete picture of plants’ sensibility to stress. To our knowledge, available drought studies examining the transcriptomic responses of tomato plants focused on gene expressions at one single WD intensity15,16,17,18,19,20, used a subjective definition of ‘mild’ WD without proper testing the actual stress levels in the plant15,19, withheld water for plants until a certain broad physiological threshold was reached (e.g. wilting)17,20, or, very rarely, by using physiological thresholds like stomatal closure16 or leaf fluorescence16,18. Unfortunately, these published studies are not comparable to each other, because only a few measured the leaf relative water content15,18 and none of them measured the leaf water potential. Consequently, we cannot assess the real stress status of plants used in these studies and understand the linear sequence of events during WD. Other publications used a completely different experimental design, via the use of osmotic stress or dehydration (leaf detachment) as a substitute for diminishing soil water availability21,22. A last group of studies23,24,25 investigated the physio-anatomical drivers of xylem embolism development without a molecular component.
Traditionally, when plant responses to WD are studied, two main approaches are applied to investigate the sequence of events leading to drought-induced plant mortality. The first set of studies focuses on the ecophysiological and/or anatomical approach, looking at modifications of the long-distance water transport pathway and/or changes in xylem anatomical traits imposed by different levels of soil water content26,27,28,29,30. A second line of publications is molecular-oriented and investigates the molecular signaling pathways and finer-scale changes in the gene, protein, and chemical chain of reactions during WD31,32,33,34. Surprisingly, these two scientific disciplines are typically separated from each other and focus on different plant groups: the hydraulic-anatomical approach deals almost exclusively with tree species, studying the sequence of events triggering the forest decline35,36, while the molecular approach mainly focuses on non-woody model species (one above all, Arabidopsis thaliana) subjected to induced stress under controlled growth chamber conditions37,38,39. Cross-pollination between the two disciplines is only sporadic, mostly limited to the quantification of the expression of key genes during the ecophysiological monitoring of species subjected to WD40,41, or constrained to the measurements of leaf water potential (Ψleaf) and gas exchange when investigating gene regulatory pathways imposed by WD16,42. Finally, only a few studies performed ecophysiological investigations in plants over- or underexpressing selected genes23,43. Consequently, achieving a deeper comprehension of plant responses to WD demands a more integrated approach, wherein the comparison of gene expression at specific time intervals during WD - based on ecophysiological monitoring - is essential.
Here, we performed a holistic drought study on the leaves of tomato plants, addressing the knowledge gaps by coupling ecophysiology and molecular biology across increasing WD intensities. We outlined three key objectives: firstly, we coupled precise ecophysiological monitoring (water potential, gas exchange, stomatal and xylem anatomy, xylem embolism spread, turgor loss point) of fully developed leaves in tomato plants with quantification of gene expression in leaves at four WD intensities: (1) partial stomatal closure, (2) full stomatal closure, (3) leaf wilting and the start of embolism formation, and (4) the point where embolism starts to exponentially increase corresponding to 12% loss of hydraulic conductivity in the main veins. Secondly, we identified and compared the gene expression at these four physiological thresholds. Thirdly, we focused on a leaf developmental stage-specific response, providing a putative transcriptomic basis for the higher resilience of younger tomato leaves to WD.
Results and discussion
Ecophysiological responses of tomato leaves to increasing water deficit
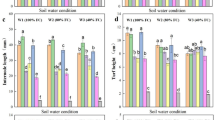
We monitored ecophysiological changes in plants subjected to a 10d period of water deprivation, while simultaneously taking samples for gene expression analyses at specific time points determined by the monitoring (Fig. 1). Under well-watered (WW) conditions where the leaf water potential (Ψleaf) was above − 0.4 MPa, the plants showed a leaflet stomatal conductance (gs) of 392 ± 18 (average ± SE) mmol H2O m− 2 s− 1 and a CO2 assimilation (A) of 8.9 ± 0.4 µmol m− 2 s− 1. These leaflets showed 154 ± 5 stomata mm− 2 with a length of 18.35 ± 0.2 μm at the abaxial side and an intervessel pit membrane thickness of 0.42 ± 0.01 μm. As the soil water content decreased, Ψleaf declined as well (Fig. 1A) and stomata closed (Fig. 1B) at Ψleaf=-0.7 MPa. The turgor loss point in leaves (ΨTLP, the key physiological indicator of a plant’s ability to maintain cell turgidity in leaves, defined when the pressure in- and outside the cells is equal) was identified at -1 ± 0.04 MPa (purple line in Fig. 1B). At a later stage of WD, air embolism appeared and spread in the leaf veins, causing 12% increase of embolized pixels (12% PEP, or P12) at -1.41 ± 0.1 MPa, while the average P50 and P88 were detected at -1.75 ± 0.1 and − 2.08 ± 0.14 MPa, respectively (Fig. 1B). The stomatal safety margin (i.e., the xylem pressure range between stomatal closure and the P50) corresponded to 1.05 MPa.
Sampling strategy and physiological thresholds in 50-day-old S. lycopersicum cv. MoneyMaker plants. (A) Leaf water potential (Ψleaf) variation over time for well-watered (WW, green circles) and water-deficit (WD, yellow circles) plants during the 10d drought experiment. The four sampling points for RNA extraction (at 4, 5, 8, and 10 days after water deficit, d-WD) are indicated with black vertical lines. Symbols represent averages ± SE (n = 3–6 per sampling point). (B) Stomatal safety margin during Ψleaf decline, defined by the difference between the best-fitted blue curve for stomata dynamics (gs, n = 33), and the red vulnerability curve (average ± SE) based on leaf percentage of embolized pixels (leaf PEP, n = 8). The purple vertical line indicates the turgor loss point (ΨTLP), and the black vertical lines indicate the four sampling dates for RNA extraction.
To elucidate the molecular processes underlying the observed physiological changes, we extracted RNA at distinct physiological stages for a comparative transcriptomic analysis through RNA sequencing (Figs. 1 and 2). To this end, we sampled comparable leaflets from mature leaves for the WW condition and at 4, 5, 8, and 10 days after water deficit (d-WD). These sampling dates corresponded to precise physiological thresholds: at 4d-WD the stomata started to close (gs10, Ψleaf=-0.41 MPa), at 5d-WD the stomata were completely closed (gs90, Ψleaf=-0.73 MPa), at 8d-WD the leaves appeared wilted and embolism started to develop in the xylem (4–6% PEP, Ψleaf=-1.3 MPa), and at 10d-DW, P12 was reached (Ψleaf=-1.43 MPa). In addition, at 10d-WD, we also sampled young developing leaves closer to the stem apex that showed no wilting despite being under a lower Ψleaf compared to mature leaves on the same plant (Ψyoungleaf=-1.61 versus Ψmatureleaf=-1.43 MPa on average).
Transcriptomic profiling of water deficit (WD) and well-watered (WW) tomato plants. (A) Multidimensional scaling plot visualizing (dis)similarity between WD and WW samples. Symbols represent biological replicates, colors represent the different treatments (yellow for WD, green for WW conditions), and different shapes represent the different sampling points (circle for 4d-WD, square for 5d-WD, rhombus for 8d-WD, triangle for 10d-WD, and plus signs for young leaves at 10d-WD). (B) Expression heatmap and hierarchical clustering of the top 500 most variable genes. Each column corresponds to one replicate: WW or WD, mature (M) or young (Y) leaves, and different clusters are separated by white lines. All biological replicates cluster to their respective time points and treatments. Color is determined by z-score, ranging from − 3 (blue) to 3 (red). (C) Volcano plots showing differentially expressed genes between WW and WD conditions in mature leaves for every time point. Differential expression is defined as having a log2 fold change over WW conditions of at least 1, and a Benjamini-Hochberg FDR-adjusted p-value below 0.05.
Transcriptomic profiling during increasing water deficit
Over 1 billion paired clean reads were successfully mapped across all samples to the SL4.00 genome release, with 93.35–95.47% of counted fragments uniquely mapped to genes according to the ITAG4.1 gene annotation release (Suppl. Table S1). A multidimensional scaling plot shows that all the WW samples cluster close together, showing comparable transcriptomic profiles and indicating that our control samples are similar (Fig. 2A). In contrast, a clear progressive separation over Dimension 1 (representing 61% of the observed variance) is seen for WD samples, correlated with progressively increasing WD (Fig. 2A). Similarly, hierarchical clustering analysis of the top 500 most variable genes across all samples clustered all replicates to their respective time points and conditions (Fig. 2B), highlighting the low variability between biological replicates and the overall good quality of the sampling.
We compared gene expression in the well-watered and drought-stressed plants for each time point. In total, we found 7179 genes that were differentially expressed (DE) in at least one time point (Suppl. Table S2). At 4 days after water deficit (4d-WD at 10% reduction in gs), a total of 379 genes were differentially expressed (232 genes were down- and 147 genes were upregulated; Fig. 2C). At 5d-WD at 90% reduction in gs (i.e., the stomata were fully closed) we found 1637 DE genes (1055 down- and 582 upregulated; Fig. 2C). At 8d-WD after the leaves had fully wilted and embolism started to develop, we found 5531 DE genes (3115 down- and 2416 upregulated; Fig. 2C). Lastly, at 10d-WD, when the vulnerability curve started to show an exponential increase in embolism events and the percentage of embolized pixels (PEP) reaches 12% (P12; Fig. 1B), we found 4637 DE genes (2921 down- and 1716 upregulated; Fig. 2C).
There was a pronounced difference in the number of pre- and post-stomatal closure DE genes (Suppl. Fig. S5). Most DE genes were unique to each time point, highlighting the particular importance of transcription for the specific WD intensity. However, 105 genes were commonly up- or down-regulated during all WD intensities, whereas a much larger portion (1011) was differentially expressed in post-stomatal closure samples (from 5d- to 10d-WD), indicating an important shift in the leaf physiology imposed by (and/or stimulating) stomatal closure. Similarly, a large fraction of DE genes (2514) is shared across the post-wilting samples, when embolism starts to develop in leaf xylem.
Next, we examined the clustering and expression patterns of genes via a weighted gene correlation network analysis (WGCNA, Fig. 3). Firstly, we restricted our analysis only to the previously identified 7179 genes that showed differential expression in at least one time point (Suppl. Table S2). To effectively characterize gene expression under increasing WD, we used a pseudo-time series, taking 4d-WD well-watered plants as basal condition, following up with WD samples for subsequent time points (4-10-d WD). Nineteen clusters of inter-correlated genes were identified (Fig. 3A). We correlated the cluster eigengenes to the physiological traits we measured on the same plants (Fig. 3B), showing their change over increasing WD. Three groups of clusters were easily detectable. The first group (clusters i-vi) showed a positive relationship between gene expression and Ψleaf, gs, and A (Fig. 3B), a negative relationship with the increase of embolism (leaf PEP, Fig. 3B), and a general downregulation during increasing WD (Fig. 3C). The second group (clusters vii-xiii) showed the opposite trend: a negative relationship between gene expression and Ψleaf, gs, and A (Fig. 3B), a positive relationship with the increase of embolism (leaf PEP, Fig. 3B), and a general upregulation during WD (Fig. 3C); the third group (clusters xiv-xix) showed an intermediate regulation compared to the two previous groups and a significant relationship with few or no recorded traits.
Weighted gene correlation network analysis (WGCNA). (A) Dendrogram denoting WGCNA results for all previously identified DE genes. The y-axis (Height) represents the distance, or dissimilarity between clusters. Nineteen distinct co-expression clusters were identified, with different colors and Roman numerals (i-xix). Gene cluster designations can be found in Suppl. Table S2. (B) Cluster-trait relationship heatmap, correlating the module eigengenes (ME) to the measured physiological traits: leaf water potential (Ψleaf), stomatal conductance (gs), CO2 assimilation (A), and loss of hydraulic conductance in leaves (leaf PEP). Correlations are plotted on a diverging color scale centered on 0 (yellow), with red denoting a positive correlation and green a negative correlation. (C) Expression z-scores for each cluster, using scaled and centered VST normalized counts over increasing water deficit from basal (WW) conditions, consisting of WW plants on the first sampling date, to 10d-WD.
Co-expression gene network analyses indicate key regulatory genes involved in the different stages of the water deficit response
To further explore the transcriptional regulation of the WD response, we prioritized identifying the most vital transcription factors (TFs) given their pivotal roles in regulating gene expression. We argued that TFs are important for two primary reasons: (1) they could exhibit the highest differential expression at a given time point and (2) they could exert a high influence on the network. To this end, we calculated centrality measures (degree and betweenness centrality) for all previously identified TFs of interest, in addition to examining differential expression per time point (Fig. 4; Suppl. Fig. S6). Two TFs in particular (Solyc01g095460.3 and Solyc05g050220.3; Suppl. Fig. S6) presented the highest number of correlations and betweenness centrality over the four sampled dates. These two TFs belong to the G-box binding factor (GBF) family and their expression increases with increasing WD (cluster viii). They are close homologs of AtGBF3, an abscisic acid (ABA)-activated Arabidopsis gene that confers increased tolerance to drought when overexpressed44,45. ABA is a critical hormone in abiotic stress signaling known to accumulate during WD stress46,47. Our data suggest that both genes play a pivotal role in the drought response of tomato leaves, likely by triggering a cascade of events related to the ABA-dependent response to WD.
Transcription factor expression and number of correlated genes over a progressive water deficit. Transcription factors are separated by which sampling time point they first break the differential expression threshold (log2 fold change > 1; padj < 0.05), focusing only on upregulated transcription factors. In the columns (from left to right) we indicated: a histogram showing the distribution of correlation strengths (weight) of correlated genes, grouping weights within 0.01 difference together; the gene cluster (see Fig. 3); the number of correlated genes from 0 (light yellow) to 1500 (green); the differential expression for each transcription factor across time points is given in log2 fold change spanning from − 8 (blue) to 8 (red) and centered on 0 (white); and the locus ID of the gene.
Subsequently, we looked for TFs that drive the WD response at the specific time points. We subdivided the previously identified TFs of interest by the time point at which they first show significant upregulation (Fig. 4). At 4d-WD, at the onset of stomatal closure (Fig. 1B), the TFs presenting the highest number of correlations (all belonging to cluster xvi) followed an ABA-independent molecular pathway: Solyc03g124110.2 and Solyc03g026280.3 are two genes presenting a strong difference in expression compared to control plants (4.21 and 2.65 LFC, respectively); they are close homologs of Arabidopsis AtDREB1 (one of the key TFs driving the ABA-independent response to drought48,49). Alongside, Solyc04g078640.3, Solyc12g009240.1, Solyc03g093540.1, and Solyc03g093550.1 belong to the Ethylene Response Factor (ERF) family, members of which are often, but not always, repressed by ABA50. It has been shown that hydraulic signals (i.e., changes in water potential) regulate stomatal response to WD before ABA accumulates in the stressed leaves imposing a more stable stomatal closure51,52,53. Our data corroborates this, as the ABA-independent TFs were driving the co-expression network in tomato leaflets that showed a reduction of 10% in stomatal conductance and a Ψleaf=-0.41 MPa.
At 5d-WD, the time point of full stomata closure, the leaves were not yet wilting, and the onset of embolism did not occur in the leaf xylem (Fig. 1). The TFs presenting the highest number of correlations belong to cluster viii (i.e., increasing linearly with WD intensity) and are mostly related to the ABA-dependent pathway (Fig. 4). More specifically, they included Solyc01g095460.3 and Solyc05g050220.3, close homologs of AtGBF3 described above, and Solyc04g078840.3 (AREB1, ABA-responsive element binding protein 1), one of the most important TFs in the ABA-dependent response to drought in many plant species54,55,56,57,58. We also retrieved Solyc12g044390.3, Solyc06g066540.1, and Solyc03g120840.3, close homologs of AtDREB3, known to be involved in abiotic stress tolerance59. In addition to the genes involved in the ABA-dependent response to drought, we found two genes involved in the biosynthesis of antioxidant molecules: Solyc10g079150.3 and Solyc10g081840.3 are two SlNF-YA TFs involved in flavonol biosynthesis60. Furthermore, Solyc07g040680.3 and Solyc06g053960.3 encoding the two heat shock factors SlHsfA2 and SlHsfA6b, respectively61, were highly correlated as well. In summary, this shift in expression occurring at 5d-WD likely indicates an accumulation of ABA in the leaves of (rather sensitive) tomato plants that become more drought-stressed. The flavonol biosynthesis TFs point to the beginning of the antioxidant response, probably limited to the most sensitive leaf mesophyll tissues62, and the upregulation of heat shock factors indicates an increase in leaf temperature resulting from fully closed stomata that strongly reduce water loss and leaf cooling63,64.
At 8d-WD, the leaves were wilting, and embolism events started to develop in the leaf xylem (4–6% PEP). We observed an increase in the number of correlations and betweenness centrality of additional heat-shock responsive TFs Solyc03g097120.3, Solyc04g016000.3, and Solyc06g050520.3 indicating increasing leaf temperature and the pronounced role of heat shock factors in severe WD65,66 (Fig. 4). Three WRKY TFs were also strongly upregulated, probably related to the antioxidant response (see SlWRKY01, SlWRKY24, and SlWRKY35 below). In addition, Solyc01g100460.3, Solyc01g104650.3, and Solyc02g092090.3 belonging to the bZIP family were highly correlated and probably involved in C and N metabolism reprogramming67. Aside from the analysis of TFs, we observed that genes encoding enzymes for the biosynthesis of trehalose (Solyc02g071590.3, Solyc07g006500.3) and proline (Solyc06g019170.3, Solyc02g068640.3) were upregulated from 8d-WD onwards. These are considered two of the most important osmoprotectants, meaning that they regulate the internal cell pressure against the dehydrating apoplast68,69. This indicates that, in these later stages of stressful WD conditions, retaining residual water as much as possible is vital for leaf mesophyll cells. Interestingly, enzymes involved in proline degradation (Solyc02g089620.3; Solyc02g089630.3) were already downregulated at 4d-WD, demonstrating that the osmoprotectant response is one of the earliest molecular responses to WD in tomato plants.
At 10d-WD, embolism in mature leaves reached P12, corresponding to the point after which embolism started to spread exponentially in the leaf veins. Fewer TFs were upregulated, among which Solyc02g090310.1 and Solyc05g010516.1 presented the highest number of correlations (Fig. 4). Only one of them is well described (Solyc02g090310.1); its Arabidopsis homolog, AtDOF1 (Dof zinc finger protein1), is activated by oxidative stress70, indicating the strong presence of reactive oxygen species (ROS) molecules in highly stressed leaves.
The role and timing of four transcription factor families during water deficit in mature tomato leaves
As an alternative approach to disentangle the transcriptome profiles across the different time points during our tomato drought experiment, we identified genes from four key TF families that presented a significantly higher or lower expression than in control plants (Table 1).
First, the ABF/AREB (ABRE BINDING FACTOR/ABA-RESPONSIVE ELEMENT BINDING PROTEIN) genes encode important transcription factors involved in ABA signaling; they are generally upregulated in the response of plants to abiotic stresses56,57,58. We found three genes of this family to be upregulated during WD, confirming their role in the response of tomato to drought (Table 1).
Second, the WRKY family of transcription factors has been described to be involved in plants’ growth development and response to a wide range of abiotic and biotic stresses71,72,73. In our experiment, the majority of the differentially regulated WRKY genes were found to be downregulated during WD (i.e. 27 genes belonging to clusters i to iv, Table 1), indicating a shift towards WRKY deactivation, possibly reducing cell metabolic activities. For example, SlWRKY81 has been described as a negative regulator of drought tolerance by maintaining open stomata and repressing proline biosynthesis during water shortage74,75. In our experiment, the expression of SlWRKY81 was strongly downregulated at the highest intensities of WD. Conversely, three other WRKY genes (SlWRKY01, SlWRKY24, and SlWRKY35) previously described to play a role in biotic stress response and carotenoid biosynthesis in tomato76,77, were upregulated during increasing WD. Our results also suggest that these three TFs might be involved in triggering and/or intensifying the antioxidant response of leaves during drought. Four other genes in this family (SlWRKY11, SlWRKY52, SlWRKY54, and SlWRKY55) exhibited an intermediate regulation, being upregulated at 5d-WD (at stomatal closure) and downregulated at 8d-WD (leaf wilting and onset of xylem embolism) and 10d-WD (at P12; cluster xvi, Table 1). SlWRKY52 was previously described to have a role during drought stress78, whereby its overexpression generally increased tolerance to drought and osmotic stress in tomato plants. Our results indicate that SlWRKY52 is activated only during stomatal closure and not at higher WD intensities, confirming the hypothesis of Jia et al.78 of its potential role in ABA biosynthesis and signal transmission.
Third, the NAC (NO APICAL MERISTEM, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR, CUP-SHAPED COTYLEDON) transcription factor family included 7 genes that were downregulated with increasing WD intensity, while 15 genes were upregulated during WD (Table 1). NAC family transcription factors are one of the largest plant-specific families, and some members are well-known to be involved in abiotic stress responses79,80. Among the upregulated genes, SlNAC63/RD26 and SlNAC90 were previously described to be upregulated during WD16, confirming our results showing an expression that increased linearly with increasing stress. Interestingly, three other NAC genes (SlNAC64, SlNAC78, SlNAC88) were upregulated only at 5d-WD (Table 1) but downregulated at the other WD intensities, suggesting a specific role during the earlier WD stages at stomatal closure.
Fourth, the nuclear transcription factor Y (NF-Y) family encodes genes that operate in a multiprotein complex consisting of three subunits (NF-YA, NF-YB, and NF-YC). While these TFs are mostly described for their role in flavonol biosynthesis during fruit ripening in tomato60, they were observed to be involved in growth and flower development in multiple plant species81. Interestingly, SlNF-YA9, SlNF-YB8, SlNF-YC1, and SlNF-YC9 showed a progressive increase along the increasing WD stages (Table 1), indicating a possible role of these TFs in the antioxidant response in stressed leaves. Likewise, we also found SlNF-YA10, which was shown to be involved in increased tolerance to oxidative stress in leaves and fruits of tomato plants82, to be upregulated during increasing WD. Finally, two more NF-Y TFs (SlNF-YC04 and SlNF-YC10) were upregulated exclusively at 8d-WD, the time point of the onset of xylem embolism in the leaves.
Comparative transcriptomic analyses explain stronger water deficit tolerance of young compared to mature leaves
Young developing leaves had a more negative water potential than mature and fully expanded leaves, both in WW (Ψleaf=-0.5 ± 0.05 MPa versus − 0.44 ± 0.07 MPa), and in WD (Ψleaf=-1.6 ± 0.04 MPa versus − 1.4 ± 0.07 MPa) conditions. This observation is consistent with the cohesion-tension theory, where an increasingly negative water potential pulls water from the bottom to the top of the plants83. Interestingly, despite this more negative Ψleaf, the young developing leaves at 10d-WD did not show any sign of wilting and/or yellowing, while the mature leaves were well below their wilting point (Suppl. Fig. S1). From an evolutionary standpoint, natural selection may lead to prioritizing the survival of younger leaves over older ones as older leaves are often less efficient in photosynthesis due to age-related decline84,85. This idea is also corroborated by studies highlighting the allocation of resources to younger tissues during stress- and season-related leaf senescence86,87. Likewise, it has been shown that leaves produced later in the season have an increased WD tolerance compared to leaves produced in the early spring in several woody species88,89.
We aimed to elucidate the mechanisms behind this apparent resilience of younger leaves by comparing the transcriptomic profiles between young and mature leaves under WW and 10d-WD conditions in the same plants. We identified 360 differentially regulated genes (i.e., genes with an LFC (log2 fold change) above 2, or below − 2, and an FDR-adjusted p-value below 0.05) from the interaction between differentially expressed genes between leaf age and watering regime (Suppl. Table S3). We grouped these genes into clusters of similar patterns of expression via K-means clustering. We identified seven distinct gene expression clusters (Fig. 5), which potentially delineate a functional separation in the response to WD. To avoid confusion with our previous clustering related to the WD response in mature leaves only, we named these clusters 1 to 7. To further dive into the potential biological significance of these expression patterns, we performed a Gene Ontology (GO)-enrichment analysis, a manual examination of the AHRD gene descriptions, as well as an examination of genes commonly found to be involved in the WD response4,90,91. Below, we limit our discussion to the potential role of the genes in cluster 1 (upregulation of 26 genes in young leaves at 10d-WD) and cluster 5 (downregulation of 97 genes in young leaves at 10d-WD; Fig. 5).
Comparative transcriptomic analysis of young and mature leaves under well-watered and water-deficit conditions. (A) Heatmap showing z-scores of differentially regulated genes between young and mature leaves under well-watered (WW) and water deficit (WD) conditions (10d-WD). Z-scores are represented by a diverging color scale from low (blue) to high (red), centered on 0 (light yellow). Clusters were determined by K-means clustering and cluster assignments (numbered 1-7) are given on the left-most column. Gene cluster assignments are given in Suppl. Table S3. (B) Line plots detailing average (± SE) gene expression in each cluster for young and mature leaves under WW and WD conditions. Values are given in variance stabilized transformed (VST) counts. The average gene expression for mature leaves is shown in yellow, and for young leaves in blue.
Cluster 1 contains genes that show an upregulation in WD-young leaves, while they are neither responsive in mature leaves (WW and WD) nor in WW-young leaves (Fig. 5). Thus, this cluster could potentially aid in conferring a WD tolerance specifically to younger leaves. For instance, we found that cluster 1 is enriched for genes involved in cell wall biosynthesis (Suppl. Fig. S7) with multiple xyloglucan endotransglucosylase enzymes (XTH; Solyc03g093120.5, Solyc03g093080.3, Solyc03g093130.3). XTHs catalyze the cleavage and (often) re-ligation of xyloglucan molecules to cellulose and can have dual roles in cell wall rigidity92,93. In Arabidopsis, constitutive overexpression of hot pepper (Capsicum annuum) CaXTH3 enhanced tolerance to salinity and WD, likely through increased cross-linking of xyloglucan molecules, thereby decreasing plasticity and elongation of the cell walls94,95. It is plausible that increased cell wall deposition and rigidity in younger leaves contribute to the maintenance of turgor pressure in smaller cells, at the expense of cell division and expansion (i.e., ceased leaf growth). This hypothesis is corroborated by the increased expression in young leaves of cellulose synthase (Solyc02g072240.3) and fasciclin-like arabinogalactan protein (Solyc11g069250.2) in cluster 4, and the decreased expression of expansins (Solyc09g018020.3, Solyc06g049050.3) in cluster 5.
Cluster 1 also contains a callose synthase (Solyc11g005980.3). Callose deposition is intricately connected to plasmodesmata permeability and is tightly controlled by two antagonistic enzymes: the synthesizing callose synthases/glucan synthase-like protein (CS/GSL), and breakdown β-1,3-glucanases (BGs)96. Interestingly, one of the closest homologs of Solyc11g005980.3 in Arabidopsis (AtGSL12/AtCalS3) has been connected to callose deposition in sieve elements in the phloem, where increased activity led to clogging of the plasmodesmata pores and impaired phloem loading96,97. In our dataset, one BG (Solyc01g060020.4) is strongly downregulated during WD uniquely in young leaves. The increased expression of the callose synthase gene Solyc11g005980.3, together with the decreased expression of the callose breakdown gene Solyc01g060020.4 in young tomato leaves during WD conditions suggests a combined approach to stimulate callose deposition in the phloem thereby blocking outward sugar transport. Soluble sugar accumulation during WD has been related to an increased osmoprotective power delaying leaf wilting98, as well as a ready source of energy during the antioxidant response99. Rosa et al.100 highlighted a whole chain of gene up- and down-regulations coordinated by soluble sugars during abiotic stresses. Our transcriptomic profile also suggests that young leaves accumulate sugars, which in turn might be related to (if not coordinating) the accelerated response to dehydration in these developing leaves.
Cluster 5 contains genes that show a much higher expression in young leaves under WW conditions, while they are downregulated to the same level as in mature leaves during WD (Fig. 5). This cluster may contain genes that are “shut off” during WD in younger leaves. Aside from the previously mentioned expansins, this cluster is enriched with nine genes encoding for chlorophyll a/b binding proteins (Solyc08g067330.1, Solyc02g070980.1, Solyc06g069730.3, Solyc02g070970.1, Solyc02g070990.1, Solyc02g071000.1, Solyc02g070950.1, and Solyc02g071010.1). The increased expression of the chlorophyll a/b binding protein genes under WW conditions is indicative of leaves undergoing active growth, as actively proliferating cells build their photosynthetic machinery101. The reduction in expression of these genes during WD coincided with the shifting in cell wall structure (and consequently growth cessation) highlighted in cluster 1, indicating that photosynthesis as well is affected at this WD intensity. Moreover, even under well-watered conditions, the photosynthetic apparatus is one of the major producers of ROS, which is exacerbated by abiotic stress102,103. Therefore, the capacity of young leaves to more effectively reduce photosynthetic activity compared to older leaves may also help to reduce the oxidative pressure under WD.
We also found that young leaves maintain an overall higher presence of negative regulators of peptidase activity under WD. Two are upregulated uniquely in young leaves under WD (cluster 1; Solyc09g083435.1, Solyc03g098795.1) and three remain highly expressed in young leaves, while they are downregulated in mature leaves (cluster 2), namely one cathepsin D inhibitor protein (Solyc03g098790.3), and two protease inhibitors (Solyc09g083435.1, Solyc03g098795.1). In addition, eleven prote(in)ase inhibitors maintained a significantly higher expression only in WD-young leaves, compared to just three which maintained a higher expression in WD-mature leaves. Proteases are incredibly varied, and their functions may be both detrimental and beneficial for acquiring tolerance to WD104,105,106,107,108. The increased presence of prote(in)ase inhibitors activity in young leaves under WD could denote decreased protease activity. Although increased protease activity may aid in maintaining a functional protein/enzyme pool in the face of oxidative stress by increasing protein turnover, having a high protein turnover is resource-intensive and may not be sustainable in a prolonged drought. Considering the time point of measurement (10d-WD) it would be plausible that there is a focus on maintaining current proteins, specifically in young leaves.
Conclusions
By disentangling gene expression profiles at precise physiological thresholds during increasing water deficit in mature leaves, we identified key regulatory genes and main TF families that play a role in the drought response of tomato leaves. We highlighted the activation of ABA-independent TFs during the lowest WD intensities (at 4d-WD with partial stomatal closure, i.e. 10% reduction in stomatal conductance, gs), followed by a high transcription of ABA-dependent TFs one day later (5d-WD, full stomatal closure, i.e. 90% reduction in gs). At stronger WD intensities after 8 days, when embolism started to develop in the leaves (4–6% PEP), we observed an activation of HEAT SHOCK FACTOR genes and a general antioxidant response. Finally, at the strongest intensities after 10 days of WD, when embolism spread is about to exponentially increase (P12), TFs related to oxidative stress were highly upregulated. When transcriptome profiles of young versus mature leaves are compared after 10d-WD in the same tomato plants, we can generalize that the stronger resilience of younger leaves to WD results from a shift in resource allocation from cell proliferation and expansion toward homeostasis maintenance. Compared to mature leaves, young leaves showed increased expression of genes altering the cell wall structure, like XTHs, cellulose synthase, and fasciclin-like arabinogalactan genes, which possibly create smaller cells with more rigid cell walls that can sustain stronger turgor pressures. Moreover, a potential preferential callose deposition at the phloem sieve elements could minimize water, sugar, and nutrient loss from these young leaves, allowing them to maintain osmoprotectant capacity and energy storage. Finally, by downregulating chlorophyll a/b binding protein genes, young leaves might reduce photosynthesis-related ROS production. This combined ecophysiological-molecular dataset in tomato leaves illustrates how gene regulatory pathways and ecophysiological thresholds are tightly intertwined. We highlighted the importance of integrating complementary disciplines to obtain a better mechanistic understanding of how molecular and ecophysiological responses are intertwined. Our results can offer valuable insights for tomato breeders on how to accurately assess drought stress levels in tomato plants using both molecular and ecophysiological monitoring. Moreover, we offer a set of essential drought-induced genes that can be targeted to increase drought resilience in tomato plants. We anticipate that these outcomes will serve future studies aiming to integrate both disciplines more thoroughly, which will undoubtedly shed new light on how natural and agricultural selection has impacted the underlying mechanisms of drought response across the plant kingdom.
Materials and methods
Plant material, sampling, and water deficit experimental design
Tomato (Solanum lycopersicum cv. MoneyMaker) plants were grown in multiple batches from 2020 to 2022 following the details described in this paragraph (see below): two batches of 4 plants each were sown in September 2020 and January 2022 to follow leaf embolism vulnerability and intervessel pit membrane thickness, one batch of 21 plants was sown in January 2022 to monitor gas exchange decline during dehydration and ΨTLP, a final batch of 24 plants (12 controls and 12 WD) was sown in September 2022 for RNA-Seq sampling, Ψleaf, stomatal anatomy, and gas exchange measurements. Seeds were germinated on full-strength Murashige-Skoog media containing 3% sucrose (MS 3%). After three weeks, plants with the most developed hypocotyls were transferred into 3 L pots in a mixture of potting soil (basis biomix, Lensli® substrates, Bleiswijk, The Netherlands), vermiculite and sand in a ratio of 25:8:2, along with three tablespoons of osmocote fertilizer (ICL Growing Solutions, Geldermal, The Netherlands). At the Institute of Biology Leiden (Leiden University), all potted plants were placed in the same growth chamber at 24 °C and 70% relative humidity, with a daily 16-h light and 8-h night cycle at 200 µmol m− 2 s− 1 photosynthetic active radiation and watered every other day until the beginning of the water deficit experiment. For the RNA-Seq sampling, we divided 50-day-old plants into two groups: a well-watered (WW) control group and a water deficit (WD) group that was fully deprived of water from this point forward. To safeguard against potential plant harm that could impact gene expression analysis while monitoring WD intensity, leaf water potential (Ψleaf) was assessed every other day in a subgroup of plants (three WW and three WD individuals). This was done until critical thresholds were reached, based on already available leaf vulnerability curves linking Ψleaf with drought-induced embolism formation in leaf veins using a previous batch of plants with equal age (see section “Non-invasive optical determination of embolism vulnerability in leaves”). The monitored plants were not utilized for RNA extraction. The Ψleaf measurements were conducted using a pressure chamber (Model 1000, PMS Instrument Company, Albany, OR, USA) on a leaflet from a fully matured leaf, following the procedure recommended by Rodriguez-Dominguez et al.109. When a specific WD intensity was reached (i.e., at 4, 5, 8, and 10 days after water deficit; Fig. 1, Suppl. Fig. S1), a mature leaf from the 6-8th node from the base was used for ecophysiological monitoring, and RNA extraction as detailed in Supplementary Fig. S2.
Single-leaflet gas exchange measurements and stomata anatomy
Maximal leaf gas exchange measurements were registered between 9:00 a.m. and 2:00 p.m. on a well-exposed leaflet close to the one used for RNA extraction (Suppl. Fig. S2) using a TARGAS-1 portable photosynthesis system (PP Systems, Amesbury, MA, USA). Tomato plants (n = 33, including the ones used for RNA-Seq analyses) were grown in multiple batches from January until November 2022 as described above, and measured between 45 and 60 days after sowing. Optimal conditions of photosynthetic active radiation were set in the cuvette (1500 µmol photons m− 2 s− 1). The following parameters were recorded on each selected leaflet: stomatal conductance (gs, mmol H2O m− 2 s− 1), assimilation (A, µmol CO2 m− 2 s− 1), and water use efficiency (WUE = A gs− 1, µmol CO2 mmol H2O− 1). The same leaflet from four well-watered plants was later stored in 70% ethanol for anatomical observations of stomata density and size. Before these stomatal measurements could take place, the samples were further dehydrated using a series of 70%, 96%, and 100% ethanol, submerged in acetone, and placed in the critical point dryer (Leica EM CPD300, Leica Microsystems, Wetzlar, Germany), mounted on scanning electron microscope (SEM) stubs (12.2 × 10 mm, Ted Pella Inc., Redding, CA, USA) using carbon adhesive Table (12 mm diameter, Electron Microscopy Sciences, Hatfield, PA, USA), and coated with a Quorum Q150T S Platinum Palladium sputter coater (Quorum Technologies, Laughton, UK). The abaxial leaf side was photographed with an SEM (JSM-6480 LV, JEOL, Tokyo, Japan) at 5 kV. Stomata were counted in six photos per sample, covering a total area of 1.84 mm2, and their length was measured on 36 random stomata per sample.
Intervessel pit membrane thickness
The thickness of the intervessel pit membranes in tomato leaves was measured in the leaflets xylem of five different plants before the determination of vulnerability to embolism, see below. We detached a leaflet from a node close to the one used for embolism detection, we collected a 1 cm-long section from the main vein and immediately fixed it in Karnovsky’s fixative. As described in Thonglim et al.43, the samples were cleaned three times in 0.1 mM cacodylate buffer, then post-fixed with 1% buffered osmium tetroxide, rinsed again with buffer solution, stained with 1% uranyl acetate, and dehydrated in a series of ethanol: 1% uranyl acetate replacement, with increasing concentration of ethanol (30, 50, 70, 96%, and twice in ≥ 99%). The samples were then infiltrated with Epon 812n (Electron Microscopy Sciences, Hatfield, UK) and placed at 60 °C for 48 h in an oven. The Epon blocks were trimmed to a thickness of 2 μm using a rotary microtome with a glass knife. Subsequently, the cross-sections with many vessel–vessel contact areas were cut into ultrathin sections of 90–95 nm using a Leica EM UC7 ultramicrotome with a diamond knife. The sections were dried and mounted on film-coated copper slot grids with Formvar coating (Agar Scientific, Stansted, UK), and post-stained with uranyl acetate and lead citrate. Ultrastructural observations of intervessel pits were performed and photographed using a JEM-1400 Plus TEM (JEOL, Tokyo, Japan) equipped with an 11-megapixel camera (Quemesa, Olympus). The photos were analyzed with ImageJ (Schneider et al., 2012), and the thickness was measured in three different spots of the membrane. We measured the thickness of six to fourteen intervessel pit membranes from five different leaflets, for a total of fifty-two membranes.
Non-invasive optical determination of embolism vulnerability in leaves
Air embolism formation and propagation in the leaf xylem were measured by adapting an optical technique110 monitoring light transmission through the leaf veins. Tomato plants (n = 8) were grown in multiple batches from September 2020 until January 2022 as described above and measured between 45 and 60 days after sowing. Plants were moved to the nearby hydraulics laboratory at Naturalis Biodiversity Center, where the soil was gently washed out without damaging the root system. One leaflet (still attached to the plant and in the same position as the one used for RNA-Seq or water potential measurements, Suppl. Fig. S2) was fixed to a scanner (Perfection V850, Seiko Epson Corporation, Suwa, Japan) and scanned every five minutes during one week of dehydration using an automating IT script developed by platform Phenobois Bordeaux (INRAE, France), while the stem water potential (Ψstem) was measured at least once a day with the pressure chamber using a bagged leaflet. The resulting images were analyzed with ImageJ software111 following instructions from http://www.opensourceov.org (last consulted November 2023). The formation of embolisms over time was determined by subtracting the differences in pixels in the 1st - to 3rd -order veins among the subsequent image scans. Background noise, mainly caused by tissue shrinkage, was removed using mild filters and manual image inspection. Vulnerability curves corresponding to the percentage of embolized pixels (PEP) as a function of Ψ were fitted for every sample based on the following equation111:
\({\text{PEP}}\,=\,{\text{1}}00/\{ {\text{1}}\,+\,{\text{exp }}[{\text{S}}/{\text{25 x }}({\text{Y}} - {{\text{P}}_{{\text{5}}0}})]\} ,\)
where P50 is the xylem pressure at which 50% of embolized pixels were observed and S is the slope of the curve at the inflection point.
Pressure-volume curves and determination of turgor loss point
We determined the leaf turgor loss point (ΨTLP) in four leaflets from two well-watered plants right after the last sampling point using the pressure-volume curve method112,113. Briefly, plants were watered at pot capacity, and the next day a leaflet from a mature fully developed leaf from the 6-8th node (comparable to the one used for RNA-Seq, Suppl. Fig. S2) was detached and equilibrated inside a sealed plastic bag. The leaf was weighed with a precision scale (Practum224-1 S, Sartorius, Göttingen, Germany), its water potential was measured with the pressure chamber, and afterward air-dehydrated in the lab. Starting from the onset of dehydration, its weight and water potential were measured two to three times per day for three days until complete dehydration. Pressure-volume relationships were constructed for every sample by plotting the inverse of leaf water potential (–1/Ψleaf) against the relative water content (RWC), and ΨTLP estimated as the point of transition between the linear and nonlinear portions of the pressure-volume relationship.
RNA extraction and library preparation
A leaflet from a mature and fully developed leaf (Suppl. Fig. S2) was taken from the 6-8th node from the base and flash frozen in liquid nitrogen. In the latest sampling date (10 d-WD), also young developing leaves were sampled from the topmost (closest to the shoot apical meristem) secondary shoots. All samples were collected between 2 p.m. and 3:30 p.m. In each case, the target leaflets from two plants were combined to form a single biological replicate, and each time point contained two to three biological replicates for the WW and WD batch. Tissues were ground into a fine powder using a Tissuelyzer (QIAGEN), ensuring the samples remained frozen. Total RNA was extracted using TRIzol® Reagent (Thermo Fisher) according to the manufacturer’s instructions. Total RNA purity was assessed using 260/280 and 260/230 nm wavelength absorption ratios, using a NanoDrop™ spectrophotometer (DeNovix DS-11 FX+). Library preparation and paired-end sequencing were performed by BGI Genomics (China). In total, 27 samples had RNA of sufficient quality and quantity to be sequenced (Suppl. Table S1).
Differential expression and co-expression analyses
Read trimming, alignment of reads to the genome, and mapping of reads to gene models were performed using CLC Genomics Workbench (Qiagen). Reads were aligned to the SL4.00 genome release, using ITAG4.1 gene annotation data (obtained from the solgenomics FTP site: https://solgenomics.net/ftp/tomato_genome/, accessed in June 2023). Automated human readable gene descriptions (AHRD) corresponding to the ITAG4.1 genome were downloaded from the Solgenomics website and added manually114. Differential expression analyses were performed in R (version 4.3.1), using the DESeq2 package115 between WD and WW samples on every sampling date. Correction for the overestimation of effect sizes for lowly expressed genes was done with a log2 fold change (LFC) shrink, as described by Zhu et al.116. Differentially expressed genes were defined as having an LFC difference of at least 1 in the WD plants compared to the control, and a Benjamini-Hochberg FDR adjusted p-value (padj; as given by DESeq2) below 0.05 (Suppl. Table S2). Differential gene regulation between developing and mature leaves was determined using the DESeq2 package, calculating differential regulation on the interaction between leaf age and treatment while controlling separately for both leaf age and treatment. Genes were defined as differentially regulated using a stringent filter (LFC > |2|; padj < 0.05, Suppl. Table S3) to account for the increased number of independent variables and reduce the chance for false positives. K-means clustering was performed on Variance Stabilizing Transformation (VST)-normalized counts, as obtained from the raw read counts using DESeq2 in R. Clustering accuracy was validated manually by plotting data grouped by clusters along its three principal components (Suppl. Fig. S3).
Weighted gene correlation network analysis (WGCNA), co-expression analysis, and identification of water deficit markers
Normalized counts were obtained via a VST from the unaltered read counts using the DESeq2 package. The weighted gene co-expression network analysis (WGCNA) was performed on differentially expressed genes using the WGCNA package in R (version 4.3.1), using a soft power threshold of 20, generating a signed network on the previously normalized counts (chosen as described in Langfelder and Horvath117; Suppl. Fig. S4). Cluster-Trait relationships (using watering regime, sampling date, position of the leaf on the plant, Ψleaf, gs, A, and leaf PEP as functional traits) were calculated from the cluster eigengenes as described in Langfelder and Horvath117. Genes encoding for transcription factors (TFs) were identified from the GO term “DNA-binding transcription factor activity”. TFs of interest were subsequently identified as those that presented an LFC higher than 4 when summed across all time points. The co-expression network was generated in Cytoscape using the CoExpNetViz app (Tzfadia et al.118), using previously identified transcription factors of interest as bait genes. Transcription factors of interest were taken as being differentially expressed in WD plants compared to WW plants (LFC > |1|; padj-value < 0.05) at any time point, while the networks were generated on all differentially expressed genes irrespective of time point. A stringent filter was applied for significant correlations (p-value ≤ 0.05), with a Pearson’s correlation (r) ≥ |0.95| used to generate the co-expression network. To identify which nodes exerted the most influence on the network - and thus would represent potential transcription factors of interest - we additionally calculated betweenness-centrality between all nodes in Cytoscape. The number of correlated genes for each transcription factor of interest was extracted from the topological overlap matrix, using a weight threshold of 0.4. To visualize the gene expression clusters, z-scores were calculated from the normalized expression values by centering each value on the mean expression and scaling it by the standard deviation of the entire dataset.
Data availability
Raw aligned and mapped counts from all samples, as well as the raw sequencing files are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE270245. Differential expression results, as well as the VST-stabilized counts as used for the WGCNA network generation are provided in the Supplementary Data.
References
Chase, J. M. & Leibold, M. A. Ecological Niches: Linking Classical and Contemporary Approaches (University of Chicago Press, 2003).
Treurnicht, M. et al. Functional traits explain the hutchinsonian niches of plant species. Glob. Ecol. Biogeogr. 29, 534–545 (2020).
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575, 109–118 (2019).
Zhang, H., Zhu, J., Gong, Z. & Zhu, J. K. Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119 (2022).
Greve, P., Gudmundsson, L. & Seneviratne, S. I. Regional scaling of annual mean precipitation and water availability with global temperature change. Earth Sys. Dyn. 9, 227–2408 (2018).
Greve, P. et al. Global assessment of trends in wetting and drying over land. Nat. Geosci. 7, 716–721 (2014).
Greve, P., Roderick, M. L. & Seneviratne, S. I. Simulated changes in aridity from the last glacial maximum to 4xCO. Environ. Res. Lett. 12, 114021 (2017).
Wartenburger, R. et al. Changes in regional climate extremes as a function of global mean temperature: an interactive plotting framework. Geosci. Model Dev. 10, 3609–3634 (2017).
Xu, C. et al. Increasing impacts of extreme droughts on vegetation productivity under climate change. Nat. Clim. Change 9, 948–953 (2019).
Pingali, P. L. & Green Revolution Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. U. S. A. 109, 12302 (2012).
Ray, D. K., Mueller, N. D., West, P. C. & Foley, J. A. Yield trends are insufficient to double global crop production by 2050. PLoS One 8, e66428 (2013).
Guerreiro, S. B., Dawson, R. J., Kilsby, C., Lewis, E. & Ford, A. Future heat-waves, droughts and floods in 571 European cities. Environ. Res. Lett. 13, 034009 (2018).
Raymond, F., Ullmann, A., Camberlin, P., Oueslati, B. & Drobinski, P. Atmospheric conditions and weather regimes associated with extreme winter dry spells over the Mediterranean basin. Clim. Dyn. 50, 4437–4453 (2018).
Lionello, P. & Scarascia, L. The relation of climate extremes with global warming in the Mediterranean region and its north versus south contrast. Reg. Envriron. Change 20, 31 (2020).
Gong, P. et al. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 61, 3563–3575 (2010).
Iovieno, P. Transcriptomic changes drive physiological responses to Progressive Drought stress and rehydration in Tomato. Front. Plant Sci. 7 (2016).
Mishra, U., Rai, A., Kumar, R., Singh, M. & Pandey, H. P. Gene expression analysis of Solanum lycopersicum and Solanum habrochaites under drought conditions. Genom. Data 9, 40–41 (2016).
Zhou, R. et al. Physiological analysis and transcriptome sequencing reveal the effects of combined cold and drought on tomato leaf. BMC Plant Biol. 19, 377 (2019).
Diouf, I. et al. Integration of QTL, transcriptome and polymorphism studies reveals candidate genes for water stress response in tomato. Genes 11, 900 (2020).
Bian, Z. et al. A transcriptome analysis revealing the new insight of green light on tomato plant growth and drought stress tolerance. Front. Plant Sci. 12, 649283 (2021).
Pirona, R. et al. Transcriptomic analysis reveals the gene regulatory networks involved in leaf and root response to osmotic stress in tomato. Front. Plant Sci. 14, 1155797 (2023).
Dong, S. et al. Transcriptomic profiling of Tomato leaves identifies novel transcription factors responding to dehydration stress. IJMS 24, 9725 (2023).
Lamarque, L. J. et al. Over-accumulation of abscisic acid in transgenic tomato plants increases the risk of hydraulic failure. Plant. Cell Environ. 43, 548–562 (2020).
Harrison Day, B. L., Carins-Murphy, M. R. & Brodribb, T. J. Reproductive water supply is prioritized during drought in tomato. Plant. Cell Environ. 45, 69–79 (2022).
Haverroth, E. J. et al. Gradients in embolism resistance within stems driven by secondary growth in herbs. Plant. Cell Environ., 1–13 (2024).
Lens, F. et al. Testing hypotheses that link wood anatomy to cavitation resistance and hydraulic conductivity in the Genus Acer. New Phytol. 190, 709–723 (2011).
Martin-StPaul, N., Delzon, S. & Cochard, H. Plant resistance to drought depends on timely stomatal closure. Ecol. Lett. 20, 1437–1447 (2017).
Corso, D. et al. Neither xylem collapse, cavitation, or changing leaf conductance drive stomatal closure in wheat. Plant. Cell Environ. 43, 854–865 (2020).
Lamarque, L. J. et al. Quantifying the grapevine xylem embolism resistance spectrum to identify varieties and regions at risk in a future dry climate. Sci. Rep. 13, 7724 (2023).
Lens, F. et al. Functional xylem characteristics associated with drought-induced embolism in angiosperms. New Phytol. 236, 2019–2036 (2022).
Hummel, I. et al. Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and Gene expression analysis. Plant Physiol. 154, 357–372 (2010).
Taiz, L., Møller, I. M., Murphy, A. & Zeiger, E. Plant Physiology and Development, 7th edition (Oxford University Press, 2024).
Thirumalaikumar, V. P. et al. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366 (2018).
Chong, L. et al. The tomato OST1-VOZ1 module regulates drought-mediated flowering. Plant Cell. 34, 2001–2018 (2022).
Choat, B. et al. Triggers of tree mortality under drought. Nature 558, 531–539 (2018).
Mantova, M., Herbette, S., Cochard, H. & Torres-Ruiz, J. M. Hydraulic failure and tree mortality: from correlation to causation. Trends Plant Sci. 27, 335–345 (2022).
Lamesch, P. et al. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012).
Singh, P. K. et al. Genomic and proteomic responses to drought stress and biotechnological interventions for enhanced drought tolerance in plants. Curr. Plant Biol. 29, 100239 (2022).
Lozano-Elena, F., Fàbregas, N. & Coleto-Alcudia, V. Caño-Delgado, A. I. Analysis of metabolic dynamics during drought stress in Arabidopsis plants. Sci. Data 9, 90 (2022).
Zhang, L. et al. Drought activates MYB41 orthologs and induces suberization of grapevine fine roots. Plant Direct. 4, e00278 (2020).
Thonglim, A. et al. Drought response in Arabidopsis displays synergistic coordination between stems and leaves. J. Exp. Bot. 74, 1004–1021 (2023).
da Silva, A. et al. Metabolic adjustment and regulation of gene expression are essential for increased resistance to severe water deficit and resilience post-stress in soybean. PeerJ 10, e13118 (2022).
Thonglim, A. et al. Intervessel pit membrane thickness best explains variation in embolism resistance amongst stems of Arabidopsis thaliana accessions. Ann. Bot. 128, 171–182 (2021).
Lu, G., Paul, A. L., McCarty, D. R. & Ferl, R. J. Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8, 847–857 (1996).
Ramegowda, V. et al. GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Sci. Rep. 7, 9148 (2017).
Waadt, R. et al. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694 (2022).
Kuromori, T., Fujita, M., Takahashi, F., Yamaguchi-Shinozaki, K. & Shinozaki, K. Inter‐tissue and inter‐organ signaling in drought stress response and phenotyping of drought tolerance. Plant J. 109, 342 (2021).
Akhtar, M. et al. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395 (2012).
Liu, S., Lv, Z., Liu, Y., Li, L. & Zhang, L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. https://doi.org/10.1590/1678-4685-gmb-2017-0229 (2018).
Müller, M. & Munné-Bosch, S. Ethylene Response factors: a Key Regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41 (2015).
Tombesi, S. et al. Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 5, 12449 (2015).
Huber, A. E., Melcher, P. J., Piñeros, M. A., Setter, T. L. & Bauerle, T. L. Signal coordination before, during and after stomatal closure in response to drought stress. New Phytol. 224, 675–688 (2019).
Abdalla, M. et al. Stomatal closure during water deficit is controlled by below-ground hydraulics. Ann. Bot. 129, 161–170 (2022).
Fujita, Y. et al. AREB1 is a transcription activator of Novel ABRE-Dependent ABA signaling that enhances Drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488 (2005).
Barbosa, E. G. G. et al. Overexpression of the ABA-Dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Rep. 31, 719–730 (2013).
Pan, X. et al. Identification of ABF/AREB gene family in tomato (Solanum lycopersicum L.) and functional analysis of ABF/AREB in response to ABA and abiotic stresses. PeerJ 11, e15310 (2023).
Fujita, Y., Fujita, M., Shinozaki, K. & Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525 (2011).
Yoshida, T. et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685 (2010).
Islam, M. S. & Wang, M-H. Expression of dehydration responsive element-binding protein-3 (DREB3) under different abiotic stresses in tomato. BMB Rep. 42, 611–616 (2009).
Li, S. et al. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 17, 36 (2016).
Huang, Y., Niu, C., Yang, C. & Jinn, T. The heat stress factor HSFA6b connects ABA signaling and ABA-Mediated heat responses. Plant Physiol. 172, 1182–1199 (2016).
Watkins, J. M., Chapman, J. M. & Muday, G. K. Abscisic Acid-Induced reactive oxygen species are modulated by flavonols to Control Stomata Aperture. Plant Physiol. 175, 1807–1825 (2017).
Bautista-Bautista, Y. et al. Genome-wide analysis of HSF genes and their role in the response to drought stress in wild and commercial Carica papaya L. genotypes. Sci. Hortic. 328, 112889 (2024).
Wang, N. et al. Heat shock factor A8a modulates flavonoid synthesis and drought tolerance. Plant. Physiol. 184, 1273–1290 (2020).
Mishra, S. K. et al. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567 (2002).
Hwang, J. E. et al. Overexpression of Arabidopsis dehydration- responsive element-binding protein 2 C confers tolerance to oxidative stress. Mol. Cells. 33, 135–140 (2012).
Dietrich, K. et al. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23, 381–395 (2011).
Iturriaga, G., Suárez, R. & Nova-Franco, B. Trehalose metabolism: from osmoprotection to signaling. Int. J. Mol. Sci. 10, 3793–3810 (2009).
Szabados, L. & Savouré, A. Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97 (2010).
Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560 (2002).
Huang, S. et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 287, 495–513 (2012).
Wani, S. H., Anand, S., Singh, B., Bohra, A. & Joshi, R. WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085 (2021).
Khoso, M. A. et al. WRKY transcription factors (TFs): molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant. Sci. 13, 1039329 (2022).
Ahammed, G. J., Li, X., Wan, H., Zhou, G. & Cheng, Y. SlWRKY81 reduces drought tolerance by attenuating proline biosynthesis in tomato. Sci. Hortic. 270, 109444 (2020a).
Ahammed, G. J. et al. Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2–mediated stomatal closure. Environ. Exp. Bot. 171, 103960 (2020b).
Moghaddam, G. A., Rezayatmand, Z., Nasr Esfahani, M. & Khozaei, M. Genetic defense analysis of tomatoes in response to early blight disease, Alternaria alternata. Plant Physiol. Biochem. 142, 500–509 (2019).
Yuan, Y. et al. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 234, 164–178 (2022).
Jia, C. et al. A group III WRKY transcription factor, SlWRKY52, positively regulates drought tolerance in tomato. Environ. Exp. Bot. 215, 105513 (2023).
Olsen, A. N., Ernst, H. A., Leggio, L. L. & Skriver, K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87 (2005).
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K. & Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. et Biophys. Acta (BBA) Gene Regul. Mech. 1819, 97–103 (2012).
Petroni, K. et al. The promiscuous life of plant nuclear factor Y transcription factors. Plant Cell 24, 4777–4792 (2012).
Chen, W. et al. A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the d-mannose/l-galactose pathway in tomato. Hortic. Res. 7, 200 (2020).
Dixon, H. H. & Joly, J. On the ascent of Sap. Philos. Trans. R. Soc. Lond. 186, 563–576 (1895).
Procházková, D. & Wilhelmová, N. Leaf senescence and activities of the antioxidant enzymes. Biol. Plant 51, 401–406 (2007).
Wang, R. H., Chang, J. C., Li, K. T., Lin, T. S. & Chang, L. S. Leaf age and light intensity affect gas exchange parameters and photosynthesis within the developing canopy of field net-house-grown papaya trees. Sci. Hortic. 165, 365–373 (2014).
Schippers, J. H. M., Schmidt, R., Wagstaff, C. & Jing, H. C. Living to die and dying to live: the Survival Strategy behind Leaf Senescence. Plant Physiol. 169, 914–930 (2015).
Berens, M. L. et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. 116, 2364–2373 (2019).
Sorek, Y., Greenstein, S. & Hochberg, U. Seasonal adjustment of leaf embolism resistance and its importance for hydraulic safety in deciduous trees. Physiol. Plant 174, e13785 (2022).
Sorek, Y. et al. An increase in xylem embolism resistance of grapevine leaves during the growing season is coordinated with stomatal regulation, turgor loss point and intervessel pit membranes. New Phytol. 229, 1955–1969 (2021).
Kido, É. A. et al. Osmoprotectant-Related Genes in Plants Under Abiotic Stress: Expression Dynamics, In Silico Genome Mapping, and Biotechnology. In Osmoprotectant-Mediated Abiotic Stress Tolerance in Plants: Recent Advances and Future Perspectives (M. A. Hossain, V. Kumar, D. J. Burritt, M. Fujita, & P. S. A. Mäkelä Eds) pp. 1–40 (Springer International Publishing, 2019).
Mishra, N. et al. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 14 (2023).
Kaku, T., Tabuchi, A., Wakabayashi, K. & Hoson, T. Xyloglucan oligosaccharides cause cell wall loosening by enhancing xyloglucan endotransglucosylase/hydrolase activity in azuki bean epicotyls. Plant Cell Physiol. 45, 77–82 (2004).
Miedes, E. et al. Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 64, 2481–2497 (2013).
Cho, S. K., Kim, J. E., Park, J. A., Eom, T. J. & Kim, W. T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 580, 3136–3144 (2006).
Hayashi, T. & Kaida, R. Functions of xyloglucan in plant cells. Mol. Plant 4, 17–24 (2011).
Wu, S. W., Kumar, R., Iswanto, A. B. B. & Kim, J. Y. Callose balancing at plasmodesmata. J. Exp. Bot. 69, 5325–5339 (2018).
Vatén, A. et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21, 1144–1155 (2011).
Ozturk, M. et al. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant 172, 1321–1335 (2021).
Sami, F., Yusuf, M., Faizan, M., Faraz, A. & Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 109, 54–61 (2016).
Rosa, M. et al. Soluble sugars: metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal. Behav. 4, 388–393 (2009).
Cackett, L., Luginbuehl, L. H., Schreier, T. B., Lopez-Juez, E. & Hibberd, J. M. Chloroplast development in green plant tissues: the interplay between light, hormone, and transcriptional regulation. New Phytol. 233, 2000–2016 (2022).
Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396 (2006).
Gill, S. S. & Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930 (2010).
Jedmowski, C. et al. Comparative analysis of sorghum bicolor proteome in response to drought stress and following recovery. Int. J. Proteom. 395905 (2014).
Cheng, L. et al. Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 16, 188 (2016).
Malefo, M. B., Mathibela, E. O., Crampton, B. G. & Makgopa, M. E. Investigating the role of Bowman-Birk serine protease inhibitor in Arabidopsis plants under drought stress. Plant Physiol. Biochem. 149, 286–293 (2020).
D’Ippólito, S., Rey-Burusco, M. F., Feingold, S. E. & Guevara, M. G. Role of proteases in the response of plants to drought. Plant Physiol. Biochem. 168, 1–9 (2021).
Moloi, S. J. & Ngara, R. The roles of plant proteases and protease inhibitors in drought response: a review. Front. Plant Sci. 14 (2023).
Rodriguez-Dominguez, C. M. et al. Leaf water potential measurements using the pressure chamber: synthetic testing of assumptions towards best practices for precision and accuracy. Plant Cell Environ. 45, 2037–2061 (2022).
Brodribb, T. J. et al. Visual quantification of embolism reveals leaf vulnerability to hydraulic failure. New Phytol. 209, 1403–1409 (2016).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Pammenter, N. W. & Van Der Willigen, C. A mathematical and statistical analysis of the curves illustrating vulnerability of xylem to cavitation. Tree Physiol. 18, 589–593 (1998).
Sack, L., Pasquet-Kok, J. & Bartlett, M. Leaf pressure-volume curve parameters. Last consulted May 1st 2023. https://prometheusprotocols.net/function/water-relations/pressure-volume-curves/leaf-pressure-volume-curve-parameters/ (2010).
Solgenomics Network. ITAG4.1_descriptions.txt [Dataset]. Last consulted May 1st 2023. https://solgenomics.net/ftp/tomato_genome/annotation/ITAG4.1_release/ (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Tzfadia, O. et al. CoExpNetViz: comparative co-expression networks construction and visualization tool. Front. Plant Sci. 6 (2016).
Jin, J. F. et al. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 21, 288 (2020).
Acknowledgements
We would like to thank Alex Bos (IBL), Jan Vink (IBL), Bertie-Joan van Hueven (Naturalis), Rob Langelaan (Naturalis), and Régis Burlett (platform Phenobois Bordeaux) for providing technical help. We want to thank Dirk Walther (MPI-MP) for verifying the bioinformatic approaches. B.M.-R. and T.A.d.W. thank the International Max Planck Research School ´Primary Metabolism and Plant Growth´ (IMPRS-PMPG; fellowship to T.A.d.W.) for support. F.L., M.L., and G.B. thank the Dutch Research Council for funding (grant ALWOP.488).
Author information
Authors and Affiliations
Contributions
G.B., T.A.d.W., B.M.R., S.B., and F.L. designed the experiments. S.B., F.L., and B.M.-R. initiated the research and provided funding. G.B., M.L., and A.T. carried out the physiological measurements. T.A.d.W. carried out the differential expression and clustering analyses. G.B. and T.A.d.W. examined genes of interest, identified structural homologs in Arabidopsis, and performed accompanying literature research. G.B. and T.A.d.W. wrote the manuscript by accepting amendments from all authors. All authors agreed on the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bortolami, G., de Werk, T.A., Larter, M. et al. Integrating gene expression analysis and ecophysiological responses to water deficit in leaves of tomato plants. Sci Rep 14, 29024 (2024). https://doi.org/10.1038/s41598-024-80261-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80261-0