Abstract
We describe the whole-genome sequencing and comparative genomic analysis of 22 mastitis-associated NASM strains isolated from India. The mean genome size of the strains was 2.55 Mbp, with an average GC content of 32.2%. We identified 14 different sequence types (STs) among the 22 NASM strains. Of these, ST1 and ST6 of S. chromogenes were exclusively associated with bovine mastitis. Genome-wide SNP-based minimum spanning tree revealed the intricate phylogenetic relationships among NASM strains from India, categorizing them into five major clades. Interestingly, mastitis-associated strains formed separate subclades in all the NASM species studied, indicating distinct host-specific co-evolution. The study identified 32 antimicrobial resistance (AMR) genes and 53 virulence-associated genes, providing insights into the genetic factors that could contribute to the pathogenicity of NASM species. Some virulence and AMR genes were found in the predicted genomic islands, suggesting possible horizontal transfer events.
Similar content being viewed by others
Introduction
Bovine mastitis is an endemic disease affecting dairy cattle worldwide. It causes inflammation of the mammary gland, resulting in economic losses due to decreased milk production, veterinary care costs, and culling of infected animals1. Staphylococci are the most common infectious causative agents of bovine mastitis, and were historically divided into two groups. One group includes S. aureus, which is considered more pathogenic, and is a coagulase-positive staphylococcus (CoPS). The other group that is presumed to be less pathogenic, includes other staphylococcal species referred to as the coagulase-negative staphylococci (CoNS) or non-aureus staphylococci (NAS). Some coagulase-positive and coagulase-variable mastitis pathogens (e.g., S. pseudintermedius) are also included in the CoNS category2. Recently, five Staphylococcus species, i.e., S. sciuri, S. fleurettii, S. lentus, S. stepanovicii, and S. vitulinus, were reclassified into the new genus, namely, Mammaliicoccous3. Together, these organisms, referred to as non-aureus staphylococci and mammaliicocci (NASM), are the most prevalent (ranging from 9.1 to 16.6% of milk samples) agents isolated from intramammary infection (IMI) of dairy cows and the leading cause of subclinical mastitis. Recent studies have identified S. chromogenes, S. epidermidis, S. xylosus, S. vitulinus, S. simulans, and Mammaliicoccus sciuri as the leading causes of IMI in cattle. Among them, S. chromogenes, S. xylosus, and S. haemolyticusare more commonly found in milk samples4,5.
Although NASM has become a leading group of pathogens, knowledge of their virulence and antimicrobial resistance mechanisms is still limited. Biofilm formation has been identified as a crucial virulence factor for NASM, particularly in persistent IMI6, and could potentially account for their heightened antibiotic resistance. The relatively increased capacity of NASM to form biofilm, as compared to other staphylococci, poses challenges in predicting antibiotic efficacy and the likelihood of persistence and recurrence of IMI. It is therefore crucial to understand the factors governing NASM resistance, persistent infections, and recurrent episodes, emphasizing the need for comprehensive research to enhance our knowledge of these intricate dynamics in the context of mastitis7.
NASM encompasses a diverse group of species, each with varying pathogenic potentials. Therefore, it is essential to understand the role of individual species on udder health and milk production2,8. Various molecular subtyping techniques, including pulsed-field gel electrophoresis (PFGE)9, random amplification of polymorphic DNA (RAPD) analysis10, multi-locus sequence typing (MLST), and multiple-locus variable number of tandem repeats (VNTR) analysis (MLVA)11, have been used to investigate the molecular epidemiology of NASM. The whole genome sequence (WGS) analysis is expected to provide the best discriminatory power and better insights into the molecular epidemiology and the genetic determinants responsible for the pathogenicity of NASM. Comparative genomic analysis can identify phylogenetic relationships among various species of NASM and distinctions within and between species.
This study focused on the comprehensive genomic analysis of 22 NASM strains associated with bovine mastitis isolated in India. These strains were fully sequenced, followed by a comparative analysis of their genetic diversity, virulence factors, antimicrobial resistance (AMR) genes, and sequence types. The study also included a perceptive phylogeny analysis of the previously documented NASM genomes from India, which helped understand the genetic differences between the NASM strains associated with or not associated with bovine mastitis.
Methodology
Whole genome sequencing and annotation
Whole genomes of 22 strains of bovine mastitis-associated NASM were sequenced. These strains were collected between 2009 and 2019 from cows (Bos taurus) with mastitis, which included 18 strains from sub-clinical and four from clinical mastitis cases in India. The strains were isolated from three states; 18 from Karnataka, and two each from Gujarat and Meghalaya. The strains had been curated at the National Institute of Animal Biotechnology in Hyderabad and the Department of Microbiology at Karnataka Veterinary, Animal & Fisheries Sciences University in Bengaluru. For isolation, milk samples collected from clinical or subclinical mastitis had been subjected to an overnight enrichment in nutrient broth, followed by streaking on selective solid media for E. coli, staphylococci and streptococci. Staphylococci had been further subjected to catalase, coagulase and thermonuclease tests, followed by PCR for nuc and/or tuf genes, which mark the isolates as S. aureus. Those that did not belong to S. aureus species had been further subjected to biochemical testing, PCR and sequencing for the 16 S rRNAgene, and in some cases, development of new PCRs for species identification12,13,14.
The DNeasy blood and tissue kit (Qiagen) was used to isolate genomic DNA from each strain, following the manufacturer’s instructions. The resulting RNA-free DNA was then employed for library preparation and sequenced using the Illumina HiSeq platform at Macrogen, Seoul, South Korea. The process of assembly and analysis of the genome sequences involved multiple steps. First, the Trimmomatic tool was used to eliminate adapters and discard low-quality reads15. The following settings were applied: ILLUMINACLIP: TruSeq3-PE.fa:2:30:10, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:36. These parameters ensured quality trimming of the raw reads, removing adapter sequences, low-quality bases, and maintaining read length integrity. Adaptor-trimmed high-quality reads were assembled using SPAdes v 3.11.116. The quality of the read assessed in checkM analysis17. Finally, the assembled genome sequences underwent annotation using the PROKKA18. Species identification was done using the rMLST (ribosomal multilocus sequence typing) tool available at the PubMLST webserver19.
Multilocus sequence typing
Multilocus sequence typing (MLST) using respective species-specific housekeeping genes was performed using the PubMLST web server (https://pubmlst.org/)20. Allelic profiles were compared to the PubMLST database, and sequence types (STs) were identified.
Average nucleotide identity (ANI) estimation
The genome sequences were analyzed to determine their Average Nucleotide Identity (ANI) values using Jspecies (https://jspecies.ribohost.com), which included MUMmer ANIm, ANItetra, and BLAST + ANIb21. S. gallinarum STP534, S. xylosus SMQ-121, S. epidermidis ATCC 14,990, S. haemolyticus ATCC 29,970, S. hominis FDAARGOS_575, M. lentus Colony453, M. sciuri NCTC12103, S. pseudintermedius FDAARGOS_930, and S. chromogenes17 A were used as reference genomes for the respective species of NASM. Each genome was assigned a species name based on an ANI cutoff of > 95%22. A heatmap was then generated using the Jspecies data on the Clustviz webserver23.
Phylogeny analysis using genome-wide SNPs
We used kSNP v.3 to predict genome-wide SNPs, identifying SNPs without requiring genome alignment24. The k-mer value of 17 was optimal for NASM strains by the KChooser tool of the kSNP v3. Genome-wide SNPs were identified and collected in a data matrix known as the 95% majority SNP matrix. Based on the genome-wide SNPs, a phylogenetic tree was constructed for 22 NASM strains using MEGA 1125. Used with the standard method, maximum likelihood with the general reversible substitution model. Additionally, a genome-wide SNP-based phylogenetic tree was constructed for 176 genomes of NASM strains reported from India, which includes 22 strains reported in this study, using Grapetree26.
Identification of antimicrobial resistance (AMR) genes and virulence factors
The resistance gene identifier (RGI) in the comprehensive antibiotic resistance database (CARD) was used to identify the AMR genes using default parameters27. The VFanalyzer in the virulence factor database (VFDB) was used to identify the genes associated with virulence factors (VF)28. We conducted our analysis in strict mode, which inherently applies a threshold thresholds of ≥ 30% identity and ≥ 80% coverage.
Identification of prophages and genomic islands
The presence of prophages in the genome was determined by PHASTER (PHAge Search Tool—Enhanced Release) (https://phaster.ca/)29. Based on the scores, prophages were classified into three groups, i.e., intact, questionable, and incomplete, with the corresponding scores > 90, 70–90, and < 70, respectively. Genomic islands (GIs) in each genome were predicted using Island Viewer 4 (http://www.pathogenomics.sfu.ca/islandviewer/)30.
Genome availability
NASM genome sequence used in this study has been deposited in NCBI with the following accession numbers: GCA_018986335.1, GCA_018996905.1, GCA_019334185.1, GCA_018997025.1, GCA_018997125.1, GCA_018967705.1, GCA_019149065.1, GCA_019149165.1, GCA_019149045.1, GCA_019193065.1, GCA_019148995.1, GCA_019334235.1, GCA_019429675.1, GCA_019149085.1, GCA_019149255.1, GCA_019149245.1, GCA_019149205.1, GCA_019334165.1, GCA_019100515.1, GCA_019165085.1, GCA_018967945.1, GCA_018968045.1.
Results
General features of genome sequences of bovine mastitis-associated NASM strains
The genomes of 22 strains of NASM associated with bovine mastitis were sequenced using the Illumina platform. These strains belong to nine different species of Staphylococcus and Mammaliicoccus genera (Table 1). For each genome, a minimum of 100X mean sequence coverage was obtained. The draft genome assemblies were checked by the checkM tool, and the genomic features such as contigs, N50, total genome length, and GC content are shown in Table S1. Adapter removal, and the processed high-quality reads were used for de novo assembly. The draft genomes contained 24 to 150 contigs with a mean genome size of 2.55 Mbp and an average GC content of 32.2%. The rMLST method uses ribosome protein subunit (rps) gene sequences for precise taxonomic identification. Of the 22 NASM strains, 21 were identified by exact matches with 56 rps genes. S. chromogenes strain K17 was identified by exact matches with 55 rps genes. The summary of genome sequences is given in Table 2.
Identification of sequence types based on MLST
MLST schemes are available for five species of NASM, which were used in this study and can be accessed at the pubMLST server. The sequence types (STs) were determined based on the respective species-specific housekeeping genes (Table S2). M. sciuri strains were divided into ST114 (n = 2) and ST115 (n = 1). Similarly, S. chromogenes were represented by ST6 (n = 2) and ST1 (n = 2). Three different STs were identified among S. epidermidis: ST1158 (n = 2), ST1157 (n = 1), and ST924 (n = 1). ST42 (n = 2) was identified in S. haemolyticus; two other strains representing potential new STs could not be typed. One strain of S. hominis belonged to ST79.
Average nucleotide identity analysis
The ANI analysis examined the interrelationship among NASM, where species-specific clustering of strains was observed, as shown in (Fig. 1) and the pairwise average nucleotide identity given in (Table S3). The ANI threshold of 96% was used to establish species delimitation within species-specific boundaries. For instance, S. chromogenes strain K17 was identical to the strains K23, K26, and K29, while S. epidermidis strain K16.1 matched the strains K4.3, K3.2, and K60. Similarly, M. sciuri strain K117.2 shared identity with strains K14 and K9, and S. xylosus strain K19 was similar to the strains K46 and SMG. M. lentus was closer to M. sciuri strains and formed a genus-specific cluster. S. gallinarum strain clustered together with S. xylosus strains. Likewise, S. pseudintermedius strain clustered with S. chromogenes, while S. haemolyticus clustered with S. hominis.
Heatmap and dendrogram illustrating the phylogenetic relationships based on average nucleotide identity (ANI) values for 22 mastitis-associated NASM isolates. The color scale represents ANI percentage identity, ranging from 75–100%, with warmer colors (orange) indicating higher similarity and cooler colors (blue/green) representing lower similarity. Unit variance scaling was applied to enhance comparability across different strain pairings. Reference genomes, marked in red, provide comparative context.
Genome-wide SNP-based-phylogeny analysis of NASM
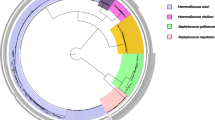
We identified 192 core SNPs and 953,968 non-core SNPs in 22 NASM strains. Based on the distribution of SNPs, a maximum likelihood phylogenetic tree using MEGA 11 was constructed with 100 bootstrap replicate as also mentioned in the branch (Fig. 2). The NASM strains grouped into five species-specific clades. S. gallinarum and S. xylosus formed a clade (I) and were divided into species-specific subclades. S. hominis and S. haemolyticus formed a clade (II) and were divided into species-specific subclades. While S. epidermidis formed a separate clade (III). S. pseudintermedius and S. chromogenes formed a clade (IV), and both species were divided into separate subclades. Similarly, Mammaliicoccus formed a separate clade (V), while M. lentus and M. sciuri formed species-specific subclades. In summary, the genome-wide SNP-based phylogeny and ANIb-based phylogeny were similar.
The genome-wide SNP-based maximum likelihood phylogenetic tree of mastitis-associated NASM strains. The phylogenetic tree was constructed using kSNP v.3, and visualized using Mega 11. The scale bar indicates 2.00 substitutions per nucleotide position and with 100 bootstrap replicates. The strain names, sequence types (STs), clinical (CL) or subclinical (SCL) mastitis, and three different states in India (GJ-Gujarat, KA-Karnataka, and ML-Meghalaya) from where the strains were isolated are indicated, respectively.
In order to further understand the inter-relationship between our strains and other NASM strains from India, a genome-wide SNP-based phylogeny analysis of 176 NASM strains (Table S4), including 22 strains from this study, was conducted. The NASM strains could be grouped into five species-specific clades (Fig. 3A). Clade A consisted of S. epidermidis, which formed a major clade with 71 strains from India, divided into two subclades. Clade B was represented by S. haemolyticus and S. hominis and divided into separate subclades. Clade C comprised S. haemolyticus, S. gallinarum, and S. xylosus. Strains belonging to the species S. haemolyticus were represented in both clades B and C. Clade D was represented by S. chromogenes and S. pseudintermedius. Clade E was represented by the Mammaliicoccus species (M. lentus and M. sciuri).
We examined the minimum spanning tree in relation to the source of isolation - whether it was from a cow or some other host. The results were interesting as all mastitis-associated NASM strains isolated from cows formed separate subclades or clusters in all clades (Fig. 3B). On the other hand, NASM strains isolated from other sources, such as humans, plants, dogs, etc. clustered together. This suggests that bovine mastitis-associated strains might have host-specific divergence and evolution.
Identification of antimicrobial resistance genes among mastitis-associated NASM strains
The 22 NASM strains contained antimicrobial resistance (AMR) genes (AMR genes) against 12 classes of antibiotics (Fig. 4). The 22 strains carried 32 AMR genes, while the analysis of all the Indian isolates revealed 57 AMR genes. The number and classes of AMR genes varied significantly among the strains. Two AMR genes, sdrM and sepA were found in all 22 strains. Of these, sdrM forms an efflux pump that confers resistance to norfloxacin and ethidium bromide, while sepA forms a multidrug efflux pump that confers resistance to disinfectants and antiseptics. The norC, which confers resistance to fluoroquinolone antibiotics, was present in all strains except for S. pseudintermedius strain B32, M. lentus, and M. sciuri species.
The dfrC, responsible for trimethoprim resistance, and the multidrug efflux pump coding gene norA, which confers resistance to fluoroquinolones as well as several antiseptics and disinfectants, were observed in all strains of S. epidermidis but absent in other species. The fusidic acid resistance gene (fusE) was detected in all strains of S. chromogenes but not in other species. The genes associated with resistance to penam drugs (mecl and mecR1) were only found in S. epidermidis strain K4.3. The methicillin-resistant gene (mecA) was present in S. epidermidis strain K4.3, S. haemolyticus strain K16.2, and S. haemolyticus strain K47. The PC1 beta-lactamase (blaZ) gene was detected in all strains of S. haemolyticus, and S. epidermidis strains K3.2 and K4.3.
The mgrA, which confers resistance to fluoroquinolone antibiotics, was present in S. haemolyticus strains A11 and A3.2, and all the strains of S. epidermidis. The salB, which confers resistance to lincosamide and class A streptogramins, was only found in M. lentus strain K169. On the other hand the homolog salC, was present in all strains of M. sciuri whereas salE was present in S. xylosus strains K19 and K46. The tetK gene, which is a tetracycline efflux protein, the qacJ gene, which confers resistance to quaternary ammonium compounds, and the SAT-4 gene, which provides resistance to nucleoside antibiotics, were only present in S. haemolyticus strains belonging to ST42. The fosBx1, which confers resistance to phosphonic acid antibiotics, was found in S. xylosus strains K19 and K46, S. gallinarum strain B13, S. epidermidis strain K16.1, and M. sciuri strains and K117.2. The fexB coding for a phenol antibiotic efflux pump was only present in S. xylosus strain SMG24. The vanT, which is a part of codes for a membrane-bound serine racemase which confers resistance to vancomycin, was present in multiple strains, except S. pseudintermedius, S. epidermidis, S. hominis, and S. gallinarum species. The vanW gene, which is part of the vancomycin operon, was identified exclusively in S. epidermidis K60. Additionally, the vanY gene, which is also part of the same operon, was present only in M. lentus K169. Furthermore, vanY was found in all strains of M. sciuri and in S. haemolyticus strains belonging to ST42, as well as in S. xylosus SMG24.
When the analysis was extended to all 176 NASM isolates from India, 57 antibiotic-resistant genes were identified in all Indian isolates as against 32 genes for our 22 strains. A complete list of AMR genes from 176 NASM strains is shown in (Table S5). And the AMR hit identity were provided in (Table S6).
Identification of virulence factors among mastitis-associated NASM strains
We identified 53 virulence factors among the 22 NASM strains associated with bovine mastitis (Fig. 5). Notably, no single virulence gene was present in all the studied strains, and each strain exhibited a unique combination of virulence factors. Adherence-related Genes, such as icaA, icaB, and icaC (intercellular adhesins), were common in S. xylosus and M. sciuri strains. Notably, the spa gene, encoding Staphylococcus protein A, was exclusive to the S. pseudintermedius strain, highlighting strain-specific adherence mechanisms. Enzyme-coding genes, lipase (lip), and thermonuclease (nuc) genes were present in all staphylococcal species but absent in Mammaliicoccus. Immune evasion genes and capsule protein genes (capB, capC) were found in most strains but absent in M. sciuri, M. lentus, and S. pseudintermedius. The adenosine synthase (adsA) gene was exclusively found in S. pseudintermedius and S. chromogenes. Toxin-related genes were strain-specific, with leukotoxin genes (lukD and lukE) found exclusively in the S. pseudintermedius strain, and cytolysin (cylR2) detected in S. xylosus, S. hominis, S. haemolyticus, and S. gallinarum strains. Only S. epidermidis K4.3 harbored genes encoding the Type VII secretion system (T7SS), esaA, esaB, esaG, essA, essB, essC, and esxA, which may contribute to its unique virulence potential. The vctC gene, responsible for iron uptake, was found in all S. xylosus strains, while the copper uptake gene (ctpV) was identified only in S. epidermidis. Genes associated with intracellular survival, such as ndk (phagosome arrest), were prevalent across Mammaliicoccus strains.
Prophage sequence-associated with NASM
We identified nine intact prophage sequences among our NASM strains. All the prophage sequences contain all the genes required for a phage life cycle, as shown in (Figure S1). Two prophage sequences, Staphylococcus phage vB Saus IMEP5 and uncultured caudovirales phage clone 10 S.1, were found in S. gallinarum. Staphylococcus phage vB Seps BE21 and Staphylococcus phage S-CoN Ph24 were found in S. epidermidis strains K60 and K16, respectively. Staphylococcus phage SAP3 was found in S. chromogenes strains K29 and K23. Similarly, S. haemolyticus strains A3.2 and A11 harbored Staphylococcus phage IME-SA4. S. xylosus contained sequences the Staphylococcus phage StB20. The presence of prophage sequences is suggestive of their potential role in the pathogenicity of NASM, however further comprehensive investigation is required.
Genomic islands of NASM
The results of the analysis of genomic islands in the 22 NASM strains are summarized in Table 3, and a representative image of the predicted genomic island is shown in (Figure S2). The number of GIs in each genome varied from 3 to 9, coding for 85 to 250 genes. These GIs mainly consisted of hypothetical protein-coding genes, transposases, and transcriptional regulators. Some known virulent factors and AMR genes were identified in a few genomic islands. For example, the fosB gene, which encodes metallothiol transferase, leading to fosfomycin resistance, was found in M. lentus. The cadC gene, responsible for cadmium resistance, transcriptional regulatory protein Yyc, which regulates the two-component system WalR/WalK regulatory protein and fosB were found in S. sciuri strains. The lip gene, coding for the virulence-related enzyme lipase, and yyc were found in S. chromogenes strains. S. epidermidis strains carried the ebh gene, coding for extracellular matrix-binding protein related to adherence, blaZ, coding for b-lactamases responsible for penicillin resistance, msr gene, which is an ABC-F subfamily ribosomal protection protein conferring resistance to erythromycin and streptogramin B class of macrolides, qac gene, which is a multidrug efflux pump resistant to fluoroquinolone antibiotics, ssaA, which is staphylococcal secretory antigen, cadC, and yyc.
S. haemolyticus strains carried farB, which is the cytoplasmic transporter protein that is part of the farAB efflux pump that confers resistance to fatty acids, arsR, coding for arsenic resistance, and blaZ, cadC, yyc, msr, and qac genes. S. hominis strains contained ebh and yyc genes. The spa, lip, msr, ssaA, and yyc genes were found in S. pseudintermedius strains. S. xylosus strains carried lip, yyc, and cadC. All S. haemolyticus strains consisted of similar islands. S. haemolyticus strains K16 and K47 had similar islands, GI-1, 2, and 3. S. haemolyticus strains A11 and A3 consisted of GI-2 in common. S. epidermidis strains K60 and K16 possessed GI-3 in common. In GI-1 of S. chromogenes strain K29 was similar to the GI-2 of S. chromogenes strain K23. The GI-1 was found in common in M. sciuri strains K117 and K91. A complete list of genomic islands from the 22 NASM strains is shown in (Table S7).
Discussion
Mastitis significantly threatens the dairy industry, causing substantial revenue losses globally. Other than S. aureus, the non-aureus Staphylococcus and Mammaliicoccus (NASM) are largely responsible for subclinical and clinical mastitis. The disease progression involves a dynamic process shaped by multiple factors, such as the host’s genetics, host as well as bacterial resistance mechanisms, host immune response, geographical influences, virulence factors (VFs) and the genetic variability of the bacterium. A comprehensive genome analysis of mastitis-associated NASM strains from diverse geographical regions could enhance our understanding of these factors, aiding in assessing pathogenic potential and infection risk, disease manifestation, and transmission dynamics.
We conducted a comparative genomic study on 22 strains of NASM that caused bovine mastitis in three states of India. We sequenced their whole-genomes and identified STs specific to each species, along with the distribution of AMR genes and virulence factors. In previous studies, an ANI threshold of < 96% was suggested for identifying NASM at the species level31. By comparing the nucleotide sequences of all the 22 strains, we found that members of the same species had ANI values consistently above 96%, indicating significant genomic similarity. On the other hand, members of different species had less than 96% ANI, indicating a clear genomic distinction between different NASM species. Our study supports the 96% ANI threshold value for species differentiation. This threshold helps understand and classify microbial diversity within this group, making it a valuable criterion for delineating species boundaries. The genome-wide SNP-based phylogeny analysis supported the ANI-based analysis, and a similar phylogenetic classification was observed.
The minimum spanning tree (MST) based on SNPs can help us understand the complex relationships among closely related strains. It can reveal patterns of co-evolution, host specialization, and potential transmission pathways. We noticed that strains associated with bovine mastitis clustered together as separate subclades in all the NASM genomes. Previous studies have reported that S. chromogenesST-1 and ST-6 strains are associated with bovine mastitis32,33. Our observation also documents that S. chromogenes ST-1 and ST-6 are associated with bovine mastitis. Additionally, we found that S. epidermidisgenotypes ST111 and ST59 are associated with bovine mastitis, which is consistent with another earlier report34. Similar sub-clusters unique to particular hosts have been observed in other pathogens35. In several European countries, S. aureus CC8 strains have been linked with bovine mastitis. Recently, we reported that S. aureusCC97 strains isolated from India were also associated with mastitis36. Further research is needed to understand the relevance of subclade-specific SNPs and their possible association with bovine mastitis to better understand their pathogenicity.
The increase in drug-resistant strains makes it challenging to treat bovine mastitis with antimicrobial intervention. In our study, we found 32 AMR genes in the 22 NASM genomes. Only one of the 22 strains had mecA and other mec-related genes. Previous studies have documented the presence of mecA-positive S. epidermidis strains in bovine milk samples and the clonal dissemination of multi-drug-resistant S. epidermidis strains carrying mecAwithin herds37. The emergence of methicillin-resistant S. epidermidis(MRSE) in cattle highlights the need for increased attention, with some researchers suggesting that animals infected with methicillin-resistant NASM should be culled38. The S. epidermidis strain K4.3 investigated in this study exhibited resistance to methicillin, classifying it as MRSE. However, the other 21 NASM strains analyzed were methicillin-sensitive staphylococcus (MSS) and did not contain mecA, mecR1, or mec1 genes in their genomes.
The blaZgene was discovered in 6 out of 22 NASM strains. This gene is responsible for the production of penicillinase, which is the primary mechanism of penicillin resistance in NASM39. Multi-drug resistant (MDR) efflux pumps were dispersed throughout the strains and were not associated with specific species or STs. The regulation of MDR efflux pumps is a complex process, requiring multiple regulators to express these elements40,41. Therefore, the presence of these efflux pumps may not necessarily translate to an AMR phenotype.
The vanG operon confers resistance to vancomycin by altering bacterial cell wall precursors, thus reducing the binding affinity of the antibiotic. Within this operon, the vanT gene encodes a serine racemase enzyme that converts L-serine to D-serine. D-serine can then substitute for D-alanine in the peptidoglycan structure, effectively lowering vancomycin’s binding ability, as the antibiotic specifically targets the D-Ala-D-Ala sequence in the cell wall. Additionally, vanW is thought to have a regulatory role, potentially aiding in stabilizing or enhancing the resistance mechanism, though its precise function remains to be fully understood. Together, vanT and vanW, along with other genes within the vanG operon, work collectively to produce a robust defense against vancomycin, particularly in species like Clostridioides difficile and Enterococcus faecalis, where the operon biological processes are still being studies42.
The vanA and vanB clusters confer glycopeptide resistance through a slightly different mechanism. They encode a D, D-carboxypeptidase enzyme known as VanY in both clusters, which cleaves the terminal D-alanine residue from peptidoglycan precursors. This cleavage prevents vancomycin from binding effectively by allowing the formation of an alternative structure, such as D-Ala-D-lactate, that the antibiotic binds poorly43. The vanM and vanF operons similarly utilize vanY to promote the formation of D-Ala-D-Lac by removing the terminal D-alanine, thereby reducing vancomycin’s effectiveness. The presence of vanY across vanM and vanF suggests a common evolutionary origin, likely from bacteria within the Bacillaceaefamily44. Collectively, these operons (vanA, vanB, vanG, vanM, and vanF) demonstrate a coordinated multi-gene response that modifies peptidoglycan precursors, allowing bacteria to evade vancomycin and continue synthesizing their cell walls despite the antibiotic’s action.
We have identified 53 virulence-associated genes among the 22 NASM genomes. Of these, 15 genes are critical for adherence and biofilm development. The icaoperon, which produce polysaccharide intercellular adhesins (PIA) and is a commonly found genetic component in biofilm formation45 stands out as a crucial player. The ica gene was present in S. xylosus and M. sciuri strain K14. The impact of icagenes is particularly noticeable within NASM species linked to the human environment46. The diversity of studies examining different variants of the icagenes poses a challenge in understanding the involvement of this gene in biofilm formation47. All of our NASM strains, excluding Mammaliicoccusspp., have genes that code for lipases. Cell-wall-associated proteins and enzymes play a significant role in the pathogenesis of staphylococci and are essential targets for drug development48. Lipases in S. aureus, known as SAL, have been identified in community-associated methicillin-resistant strains49,50.
The spa gene, a major antigen of S. aureus, was exclusively detected in S. pseudintermedius. The sspA gene was found in 12 strains, while the sspB gene was found exclusively in S. epidermidis. In S. epidermidis, the sspA and sspB genes were found to coexist, indicating a potential interplay or cooperative function. The initial player in the staphylococcal proteolytic cascade is aureolysin, a metalloprotease. This enzyme undergoes a rapid process of autocatalytic activation. Subsequently, the activated aureolysin plays a crucial role in activating the sspA serine protease, which in turn, serves as a critical activator for the sspBcysteine protease51,52. Such cascades are often pivotal in regulating various cellular processes and contribute to the pathogenicity and adaptability of members of the genus Staphylococcus. The acquisition of essential nutrients such as iron and copper is vital for the survival and virulence of Staphylococcus species, especially in iron-limited environments like host tissues where metals are tightly bound by proteins such as transferrin and lactoferrin. In non-aureus staphylococcal strains, metal uptake genes, including those for siderophores and iron transport systems, play a crucial role in enabling bacterial growth and regulating virulence-associated functions53,54. The identification of genes such as vctC for iron uptake and ctpV for copper regulation underscores the importance of metal acquisition in the pathogenic potential of NASM strains, particularly in bovine mastitis. The presence of vctC in all S. xylosus strains highlights the essential nature of iron uptake, while the strain-specific occurrence of ctpV in S. epidermidis K60 suggests unique adaptations to copper stress.
The type VII secretion system (T7SS) has been recently identified and selectively distributed in various pathogens, including Mycobacterium tuberculosis and S. aureus. The T7SS plays an essential role in the virulence of human pathogens. In the case of M. tuberculosis, the T7SS is very important for bacterial access to the host cytosol. In S. aureus, T7SS exports several virulence-associated proteins55. The presence of T7SS cluster was reported in S. lugudensis, in addition to S. aureus56. Staphylococci possess genes enabling the production of capsular polysaccharides that form a protective shield against phagocytosis by the host’s immune cells. This capsule enhances virulence and bacterial persistence, highlighting encapsulation as a crucial mechanism for evading immune detection and contributing to pathogenicity. The capB and capC genes were detected in all NASM species except Mammaliicoccus spp. and S. pseudintermedius.
Prophages are responsible for the horizontal gene transfer, which in turn contributes to virulence57,58. The prophage Staphylococcus StB20 was found in S. xylosus strain SMG24. This prophage is reported to exhibit specific proteolytic cleavages in the carboxy-terminal degradation of its tail tape measure proteins (TMP) in S. aureus59. However, the roles of prophages NASM strains are largely unknown.
Genomic islands (GIs) are horizontally transferred regions carrying specific genes that confer certain traits to bacteria. These traits include metabolic processes, pathogenicity, antibiotic resistance, and symbiosis. GIs help bacteria establish mutually beneficial relationships with eukaryotic hosts. They often carry genetic material related to virulence or adaptive traits and are commonly located near tRNA or transposase genes at one end of the island. Antibiotic-resistant genes found in GIs make them carriers for the spread of antibiotic resistance, enhancing bacterial species’ survival when exposed to antibiotics60. Coupling virulence or adaptive traits with antibiotic resistance in GIs plays a significant role in shaping bacterial populations’ resilience and adaptability to environmental challenges, especially when exposed to antibiotics. In the 22 NASM that we studied, most islands carried the two-component regulatory system, which includes the walRK gene. This gene plays a crucial role in regulating the expression of genes associated with cell wall metabolism, influencing autolysis, biofilm formation, and virulence. WalK is also involved in sensing the D-Ala-D-Ala moiety of Lipid II, serving as a signal for active cell wall synthesis. The genes yycH and yycI are co-transcribed with walRK and modulate its activity. In S. aureus, disrupting yycH and yycI genes downregulated the walRKregulon61. The importance of these regulatory components and their roles in governing cell wall-related processes vary across bacterial species. The fosB, blaZ, and several AMR genes were identified among the GIs, suggesting the possibility of transfer of these traits among the mastitis-associated pathogens.
Conclusion
Whole genome sequencing and phylogeny analysis of bovine mastitis-associated NASM strains isolated from India revealed species-specific and host-specific clustering. The study identified multiple genes responsible for antibiotic resistance and virulence factors. Certain virulence factors were found to be specific to particular species, and some were specific to particular STs. The analysis also found that some virulence and AMR genes were located in genomic islands, which suggests possible horizontal transfer events.
Data availability
NASM genome sequence used in this study has been deposited in NCBI with the following accession numbers: GCA_018986335.1, GCA_018996905.1, GCA_019334185.1, GCA_018997025.1, GCA_018997125.1, GCA_018967705.1, GCA_019149065.1, GCA_019149165.1, GCA_019149045.1, GCA_019193065.1, GCA_019148995.1, GCA_019334235.1, GCA_019429675.1, GCA_019149085.1, GCA_019149255.1, GCA_019149245.1, GCA_019149205.1, GCA_019334165.1, GCA_019100515.1, GCA_019165085.1, GCA_018967945.1, GCA_018968045.1.
References
Halasa, T., Huijps, K., Østerås, O. & Hogeveen, H. Economic effects of bovine mastitis and mastitis management: a review. Veterinary Q. 29, 18–31 (2007).
De Buck, J. et al. Non-aureus staphylococci and Bovine Udder Health: current understanding and knowledge gaps. Front. Vet. Sci. 8, 658031 (2021).
Madhaiyan, M., Wirth, J. S. & Saravanan, V. S. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. MicroBiol. 70, 5926–5936 (2020).
Valckenier, D., Piepers, S., De Visscher, A. & De Vliegher, S. The effect of intramammary infection in early lactation with non-aureus staphylococci in general and Staphylococcus chromogenes specifically on quarter milk somatic cell count and quarter milk yield. J. Dairy Sci. 103, 768–782 (2020).
Ruiz-Romero, R. A. & Vargas-Bello-Pérez, E. Non-aureus staphylococci and mammaliicocci as a cause of mastitis in domestic ruminants: current knowledge, advances, biomedical applications, and future perspectives – a systematic review. Vet. Res. Commun. 47, 1067–1084 (2023).
Simojoki, H., Hyvönen, P., Plumed Ferrer, C., Taponen, S. & Pyörälä, S. Is the biofilm formation and slime producing ability of coagulase-negative staphylococci associated with the persistence and severity of intramammary infection? Vet. Microbiol. 158, 344–352 (2012).
Srednik, M. E. et al. Biofilm formation and antimicrobial resistance genes of coagulase-negative staphylococci isolated from cows with mastitis in Argentina. FEMS Microbiology Letters fnx001 doi: (2017). https://doi.org/10.1093/femsle/fnx001
Vanderhaeghen, W. et al. Invited review: Effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J. Dairy Sci. 97, 5275–5293 (2014).
Fry, P. R. et al. Association of coagulase-negative staphylococcal species, mammary quarter milk somatic cell count, and persistence of intramammary infection in dairy cattle. J. Dairy Sci. 97, 4876–4885 (2014).
Petzer, I. M., Labuschagne, C., Phophi, L. & Karzis, J. Species identification and cow risks of non-aureus staphylococci from South African dairy herds. Onderstepoort j. vet. res. 89, (2022).
Dahyot, S. et al. Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) and Tandem repeat sequence typing (TRST), helpful tools for subtyping Staphylococcus lugdunensis. Sci. Rep. 8, 11669 (2018).
Hegde, R. et al. Incidence of subclinical mastitis and Prevalence of Major Mastitis pathogens in Organized farms and unorganized sectors. Indian J. Microbiol. 53, 315–320 (2013).
Preethirani, P. L. et al. Isolation, biochemical and molecular identification, and In-Vitro Antimicrobial Resistance patterns of Bacteria isolated from Bubaline Subclinical mastitis in South India. PLoS ONE. 10, e0142717 (2015).
Sundareshan, S. Characterization of coagulase-negative staphylococci from mastitis. Ph.D. thesis submitted to Karnataka Veterinary Animal and Fisheries Sciences University, India. (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Zakaria, M. R. et al. Genome sequence data of Mangrovimonas sp. strain CR14 isolated from mangrove forest at Tanjung Piai National Park, Malaysia. Data Brief. 30, 105658 (2020).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Larsen, M. V. et al. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539 (2014).
Aanensen, D. M. & Spratt, B. G. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 33, W728–W733 (2005).
Richter, M., Rosselló-Móra, R., Glöckner, O., Peplies, J. & F. & JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 (2016).
Colston, S. M. et al. Bioinformatic Genome Comparisons for Taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio 5, e02136–e02114 (2014).
Metsalu, T. & Vilo, J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
Gardner, S. N., Slezak, T. & Hall, B. G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878 (2015).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Zhou, Z. et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 28, 1395–1404 (2018).
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017).
Liu, B., Zheng, D., Jin, Q., Chen, L. & Yang, J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692 (2019).
Naidoo, N. & Zishiri, O. T. Comparative genomics analysis and characterization of Shiga toxin-producing Escherichia coli O157:H7 strains reveal virulence genes, resistance genes, prophages and plasmids. BMC Genom. 24, 791 (2023).
Bertelli, C. et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017).
Kim, M., Oh, H. S., Park, S. C. & Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. MicroBiol. 64, 346–351 (2014).
Huebner, R. et al. Characterization of genetic diversity and population structure within Staphylococcus chromogenes by multilocus sequence typing. PLoS ONE. 16, e0243688 (2021).
Persson Waller, K. et al. Genotypic characterization of Staphylococcus chromogenes and Staphylococcus simulans from Swedish cases of bovine subclinical mastitis. J. Dairy Sci. 106, 7991–8004 (2023).
Frey, Y., Rodriguez, J. P., Thomann, A., Schwendener, S. & Perreten, V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 96, 2247–2257 (2013).
Grillová, L. & Picardeau, M. Core Genome Multi-locus sequence typing analyses of Leptospira spp. Using the bacterial isolate genome sequence database. in Leptospira spp. (eds Koizumi, N. & Picardeau, M.) vol 2134 11–21 (Springer US, New York, NY, (2020).
Sivakumar, R. et al. Genome sequencing and comparative genomic analysis of bovine mastitis-associated Staphylococcus aureus strains from India. BMC Genom. 24, 44 (2023).
Sawant, A., Gillespie, B. & Oliver, S. Antimicrobial susceptibility of coagulase-negative Staphylococcus species isolated from bovine milk. Vet. Microbiol. 134, 73–81 (2009).
Gentilini, E. et al. Antimicrobial susceptibility of coagulase-negative Staphylococci isolated from bovine mastitis in Argentina. J. Dairy Sci. 85, 1913–1917 (2002).
Olsen, J. E., Christensen, H. & Aarestrup, F. M. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 57, 450–460 (2006).
Costa, S. S., Viveiros, M., Amaral, L. & Couto, I. Multidrug Efflux pumps in Staphylococcus aureus: an update. TOMICROJ 7, 59–71 (2013).
Antiabong, J. F., Kock, M. M., Mbelle, N. M. & Ehlers, M. M. Diversity of Multidrug Efflux genes and phenotypic evaluation of the in vitro Resistance Dynamics of Clinical Staphylococcus Aureus isolates using Methicillin; a Model β-lactam. TOMICROJ 11, 132–141 (2017).
Srinivasan, V. et al. vanG element insertions within a conserved chromosomal site conferring Vancomycin Resistance to Streptococcus agalactiae and Streptococcus anginosus. mBio 5, e01386–e01314 (2014).
Arthur, M. & Quintiliani, R. Regulation of VanA- and VanB-Type glycopeptide resistance in Enterococci. Antimicrob. Agents Chemother. 45, 375–381 (2001).
Kardos, G. et al. Phylogenetic analysis of the genes in D-Ala-D-Lactate synthesizing Glycopeptide Resistance operons: the different origins of Functional and Regulatory genes. Antibiotics 13, 573 (2024).
McKenney, D. et al. The Ica Locus of Staphylococcus epidermidis encodes production of the capsular Polysaccharide/Adhesin. Infect. Immun. 66, 4711–4720 (1998).
O’Gara, J. Ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188 (2007).
Tremblay, Y. D. N. et al. Characterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilms. J. Dairy Sci. 96, 234–246 (2013).
Hu, C., Xiong, N., Zhang, Y., Rayner, S. & Chen, S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 419, 617–620 (2012).
Cadieux, B., Vijayakumaran, V., Bernards, M. A., McGavin, M. J. & Heinrichs, D. E. Role of lipase from Community-Associated Methicillin-resistant Staphylococcus aureus strain USA300 in Hydrolyzing triglycerides into growth-inhibitory free fatty acids. J. Bacteriol. 196, 4044–4056 (2014).
Rosenstein, R. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 82, 1005–1014 (2000).
Massimi, I. et al. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease ofStaphylococcus aureus. J. Biol. Chem. 277, 41770–41777 (2002).
Nickerson, N. N., Joag, V. & McGavin, M. J. Rapid autocatalytic activation of the M4 metalloprotease aureolysin is controlled by a conserved N-terminal fungalysin‐thermolysin‐propeptide domain. Mol. Microbiol. 69, 1530–1543 (2008).
Sun, Z. et al. Determining the Genetic Characteristics of Resistance and virulence of the Epidermidis Cluster Group through Pan-genome Analysis. Front. Cell. Infect. Microbiol. 10, 274 (2020).
Ward, S. K., Hoye, E. A. & Talaat, A. M. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 (2008).
Lopez, M. et al. (ed, S.) Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. U S A 114 11223–11228 (2017).
Lebeurre, J. et al. Comparative genome analysis of Staphylococcus lugdunensis shows clonal complex-dependent diversity of the putative virulence factor, ess/Type VII locus. Front. Microbiol. 10, 2479 (2019).
Bae, T., Baba, T., Hiramatsu, K. & Schneewind, O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62, 1035–1047 (2006).
Smirnova, N. I. et al. Effect of CTXφ prophage deletion in cholera agent on expression of regulatory genes controlling virulence and biofilm formation. Russ J. Genet. 53, 302–313 (2017).
Tallent, S. M., Langston, T. B., Moran, R. G. & Christie, G. E. Transducing particles of Staphylococcus aureus Pathogenicity Island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 189, 7520–7524 (2007).
Citti, C., Baranowski, E., Dordet-Frisoni, E. & Faucher, M. Nouvel, L.-X. Genomic Islands in Mycoplasmas. Genes 11, 836 (2020).
Gajdiss, M. et al. YycH and YycI regulate expression of Staphylococcus aureus Autolysins by activation of WalRK Phosphorylation. Microorganisms 8, 870 (2020).
Acknowledgements
The MKU-RUSA, UGC-NRCBS, DST-PURSE, DST-FIST Programs of the School of Biological Sciences, Madurai Kamaraj University, are acknowledged.
Funding
This work was supported by the Department of Biotechnology (DBT), New Delhi (BT/PR11245/ADV/90/165/2014).
Author information
Authors and Affiliations
Contributions
N.R.H., J.R., and S.I. designed the experiments; M.A. and S.C. propagated and characterized the NASM strains from the various States of India; V.R. and R.S. performed WGS and analysis; V.R., S.I., J.R., and N.R.H. wrote the paper. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
We declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ramesh, V., Sivakumar, R., Annamanedi, M. et al. Genome sequencing and comparative genomic analysis of bovine mastitis-associated non-aureus staphylococci and mammaliicocci (NASM) strains from India. Sci Rep 14, 29019 (2024). https://doi.org/10.1038/s41598-024-80533-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80533-9