Abstract
The increasing demand for biotherapeutics has necessitated the evaluation of their critical quality attributes, one of which is glycosylation, an essential post-translational modification found on many biological molecules. In particular, the purification of N-glycans after their release from the proteins and derivatization is important in ensuring the removal of the deglycosylated protein, excess labelling reagents and salts for subsequent analysis. However, current methods of N-glycans purification are either expensive, laborious, time-consuming or not catered for high throughput analysis. To overcome these constraints, we developed a high throughput purification method for fluorescent derivatized N-glycans using cellulose functionalized magnetic beads (CMBs). We compared the method with two current purification methods, hydrophilic interaction chromatography solid phase extraction (HILIC-SPE) and gel filtration using human serum IgG (hsIgG) and bovine fetuin and assessed their reproducibility. The CMB method yielded highly similar glycan profiles to the two methods with very good precision. We then assessed the compatibility of the method to purify N-glycans derivatized with different fluorescent labels (RapiFluor MS, 2-aminobenzamide and procainamide). We also applied the methodology to analyse N-glycans in the biotherapeutic protein, recombinant alpha-1-antitrypsin (rAAT) which was modified with higher sialylation content. Importantly, the method successfully captured the differences in the glycan profiles between the modified and unmodified rAAT. Finally, we automated the method together with the digestion and labelling protocol onto a robotic liquid handler for high throughput glycosylation analysis. The versatility of the CMB method, together with its affordability and robustness, may provide an alternative workflow for the high throughput analysis of glycans in biotherapeutic modalities.
Similar content being viewed by others
Introduction
Glycosylation is an essential and abundant post-translational modification governed by multiple enzymes that bind carbohydrates to lipids or proteins and is critical in modulating several biological functions including their stability, folding, cell signalling, and immunogenicity1. Glycosylation also plays a key role in biotherapeutic development, where they can significantly impact their quality. For instance, glycosylation patterns can influence the pharmacokinetics (PK) and pharmacodynamics (PD) properties of biotherapeutics through their half-lives and efficacies2,3,4,5,6. Additionally, glycans can affect effector functions such as the antibody-dependent cell-mediated cytotoxicity (ADCC) activity of antibodies4,7,8,9 as well as trigger immunogenicity in humans10. Thus, to ensure the safety, functionality and efficacy of biotherapeutic drugs, glycosylation must be correctly engineered and expressed11. Indeed, glycosylation has been identified as one of the critical quality attributes (CQAs) required for biotherapeutic products, and is crucial to the development and production of biotherapeutics for their safety and batch-to-batch consistency. In particular, during biotherapeutic process optimization, there are numerous samples generated and each require characterization that can be costly. An effective and high throughput method that can accommodate large number of samples is therefore greatly desired to support the speedy turnaround and reduce cost in routine glycan profiling.
Analytical approaches for N-glycan profiling typically involve the chemical or enzymatic release of the glycans, derivatization with a fluorescent label, purification and characterization by methods such as fluorescence-based liquid chromatography (LC-FLR)12,13,14, LC-FLR mass spectrometry (LC-FLR-MS)12,15,16, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF)17,18,19 or capillary electrophoresis laser-induced fluorescence (CE-LIF)20,21. In particular, purification of the glycans after their labelling is especially important for the removal of excess labelling reagent, salts, unlabelled glycans and deglycosylated proteins that could interfere with subsequent analysis and distort the true depiction of the relative abundances of the glycans. Some purification techniques commonly in use include S cartridges22,23, gel filtration chromatography24,25, acetone precipitation12,26 and hydrophilic interaction chromatography solid phase extraction (HILIC-SPE)27,28,29. For instance, S cartridges, where glycans are adsorbed to the disc matrix for isolation, are widely employed in the biopharmaceutical and bioprocessing industries. However, they are extremely expensive compared to gel filtration chromatography and acetone precipitation procedures, though the latter methods are significantly more laborious and time consuming. A cost-efficient and high throughput approach is HILIC-SPE as it has proven to be a popular method among analytical scientists for isolating glycans due to its high separation efficiency, affordability and adaptability onto a 96-well configuration.
Over the years, there have been significant advancements in the field of glycosylation analysis such as the development of rapid glycan release and labelling kits30, utilisation of 96-well plates14,27,31, magnetic nanoparticles for cleanup20,21,32, label-free glycopeptide analysis23 and process automation14,33. Magnetic nanoparticles, in particular, have found widespread use in various applications such as immunoprecipitation34, food analysis35, nucleic acids detection36 and treatment of chronic diseases37. The use of magnetic nanoparticles in glycan purification are becoming increasingly popular due to their low cost, possibility for functionalization, high specificity, potential for high throughput analysis and process automation.
In this study, we present a novel approach of using cellulose functionalized magnetic beads (CMBs) to purify fluorescent derivatized N-glycans released from biotherapeutic proteins. We evaluated the performance of this method by comparison of its glycosylation profile and the relative abundance of the glycans obtained with two other purification strategies – HILIC-µSPE16,30 and gel filtration chromatography24,25. We also assessed the compatibility of the method to purify N-glycans labelled with different fluorescent labels and demonstrated the applicability of the CMBs to purify N-glycans released from recombinant alpha-1-antitrypsin (rAAT). Finally, we automated the labelling method together with the purification method onto a liquid handler platform, showcasing its potential as a high throughput purification and profiling method for N-glycosylation analysis in biotherapeutic modalities.
Results
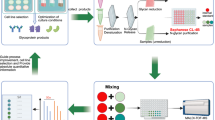
Framework of the purification workflow using CMBs
In our purification strategy, we incorporate the use of magnetic beads that were functionalized with cellulose to purify fluorescent derivatized N-glycans in the workflow shown in Fig. 1. In the first part of the workflow, N-glycans are first released from the proteins via enzymatic digestion using the enzyme, peptide N-glycosidase F (PNGase F) and labelled subsequently with a fluorescent tag. Subsequently, the CMBs are added to the solution, where the N-glycans will bind to the magnetic beads via hydrophilic interactions. In particular, cellulose was identified as an ideal functionalisation material for the magnetic beads as the presence of numerous hydroxyl groups in the polysaccharide molecule confers high polarity to the beads, thereby resulting in its strong affinity for the hydrophilic N-glycans via hydrogen bond formation. As such, the CMBs are able to separate the labelled N-glycans from the rest of the impurities such as the excess fluorescent label, salts, and deglycosylated proteins in the sample, yielding a highly purified sample of N-glycans to be collected at the end of the process. We optimized the binding time of the beads to the glycans, testing at 5 min and 10 min using N-glycans released from human serum IgG (hsIgG) and bovine fetuin, labelled with Rapifluor-MS (RFMS). While we observed high variation in relative abundances (> 20%) for some of the glycans due to co-elution and their low abundances, there were no major differences overall between the two binding times (average RSD < 6%) (see Supplementary Fig. S1). Hence, a 5 min binding time was chosen for the method. Elution of the N-glycans from the CMBs is accomplished by the addition of water that releases the N-glycans from the beads, followed by characterization through LC-FLR-MS for identification and fluorescence-based quantification of the glycans.
Schematic of the CMBs purification workflow. N-glycans are first enzymatically released from the protein and labelled with a fluorescent tag. CMBs were then added to isolate the fluorescent labelled N-glycans followed by a washing step and elution of the purified products from the beads before they are analysed by LC-FLR-MS.
Evaluation and comparison of CMBs as a purification strategy to isolate fluorescent labelled N-glycans
To evaluate the performance of CMBs as a purification method for fluorescent labelled N-glycans, we used them to characterize commercial model glycoproteins, namely hsIgG and bovine fetuin. As a comparison to other established methods, RFMS labelled N-glycans from these proteins were subjected to purification either using our proposed CMB method, HILIC 96-well µSPE elution plate, or PD MiniTrap G10, a gel filtration column. Purified fluorescent N-glycans from each method were analyzed through LC-FLR-MS, where the peaks in the LC chromatographs were identified and relative abundances of each glycan determined by integrating the area under the peak (see “Materials and methods”).
Glycosylation profiles generated by all three approaches were found to have highly similar FLR signal intensity for both proteins (Fig. 2). There were no differences in the glycans identified among the three methods (Table 1). In quantitation, HILIC-µSPE and CMBs had similar relative abundance, although the G10 method appeared to report lower relative abundances for some of the neutral glycans (A2, FA2, FA2G1, FA2BG1) but higher abundances for the sialylated species in hsIgG, suggesting there may be some selectivity involved in the method (Table 1).
The reproducibility of the methods was assessed using four replicates and the overall average relative standard deviations (RSDs) for the N-glycans identified in hsIgG and fetuin for CMBs were 2.54% and 3.37% respectively. (Table 1). In HILIC-µSPE, it was 2.06% (hsIgG) and 0.94% (fetuin) and for G10, 7.45% (hsIgG) and 1.80% (fetuin) (Table 1). Thus, we have demonstrated the feasibility of CMBs to isolate fluorescent derivatized N-glycans with good repeatability, comparable with current purification techniques.
Comparison of N-glycan profiles of hsIgG and bovine fetuin purified with three different methods. hsIgG and bovine fetuin were derivatized with the fluorescent label RFMS and purified using CMBs, HILIC 96-well µSPE and PD MiniTrap G10. The profiles generated from the three methods were highly similar for both proteins. Red triangle represents fucose, green circle represents mannose, blue square represents N-acetylglucosamine, yellow circle represents galactose and pink diamond represents sialic acid.
Compatibility of CMBs with different fluorescent labels
The fluorescent labelling of N-glycans is required for their quantitative detection via LC-FLR-MS approaches by enhancing their signal intensities and ionization efficiencies. Consequently, the chemistry of the fluorescent labels (which are mostly hydrophobic) can affect the overall hydrophilicity of the labelled glycans and influence their interactions with the purification material utilized. Thus, a purification method suitable for separating N-glycans labelled with one fluorescent tag may not be compatible with another. For example, different types of S cartridges are required for the purification of 2-aminobenzamide (2-AB) and procainamide (ProC) labelled glycans. To that end, we sought to investigate the compatibility of CMBs to purify N-glycans labelled with 2-AB and ProC in hsIgG and fetuin compared to RFMS.
We found the glycosylation profiles obtained for hsIgG between the three labels were similar albeit lateral shifts in retention time of the eluted glycans, as previously observed16 (Fig. 3). 2-AB labelled glycans were found to elute earlier than those tagged with ProC and RFMS. For fetuin, the majority of the ProC labelled glycans were observed to elute earlier compared to 2-AB and RFMS tagged glycans. In practical qualitative analysis, different elution times can be standardized to glucose units (GU) using dextran ladders and matching GU databases developed for each label38. Interestingly, the fetuin glycan profiles for the three labels were not similar, unlike in hsIgG, with the biggest differences lying in the time region eluting before Peak 3. We noted that there were around 6 additional peaks that eluted in this region for the 2-AB and ProC glycan profiles. These 6 peaks were present also in other studies that utilized 2-AB and ProC to label bovine fetuin39,40,41 and therefore was not caused by the CMBs. The absence of these peaks in the RFMS glycan profile however, could be due to the differences in labelling efficiency of the labels. To enable a fair comparison between the three labels, these peaks were not taken into account when determining the relative abundances of the glycans for fetuin. The abundance distributions between the three labels were similar for both hsIgG and fetuin with good reproducibility although the fetuin N-glycans labelled with ProC displayed greater variations among the replicates (n = 4) (Supplementary Table S1). All in all, the ability of CMBs to clean up various fluorescent derivatized N-glycans implies that the beads have the potential to be a one-stop purification approach with the flexibility to accommodate various experimental setups.
Comparison of N-glycan profiles of hsIgG and bovine fetuin derivatized with different fluorescent labels. N-glycans from hsIgG and bovine fetuin were labelled with the fluorescent tags RFMS, 2-AB or ProC, and purified with CMBs. The glycan profiles for hsIgG were similar across the three labels albeit the lateral shifts in the retention time. However, the dissimilarities in the glycan profile for fetuin could be due to the chemistry and labelling efficiencies of the labels. represents fucose, represents mannose, represents N-acetylglucosamine, represents galactose and represents sialic acid.
Applicability of CMBs to purify N-glycans released from biotherapeutic proteins
Having demonstrated the potential of CMBs to isolate fluorescent derivatized glycans, we applied our developed workflow for the use of biotherapeutics characterization, an area that widely uses glycomics as part of CQA assessment. Specifically, we characterized N-glycans from our in-house produced recombinant alpha-1-antitrypsin (rAAT) which has been modified using bacterial sialyltransferase to produce highly sialylated AAT that can potentially have better PK properties for AAT deficiency treatment due to the higher degree of sialylation5,42. Indeed, the recent advancements of such glycoengineering efforts necessitate robust analytical procedures for the characterization of unique and complex glycoforms, often for many samples. We released N-glycans from both rAAT and rAAT that has been modified with bacterial sialyltransferase, labelled them with RFMS and carried out the purification using CMBs. The three major N-glycans observed in the modified rAAT were FA2G2S2 (25.0%),FA2G2S1 (21.4%),and A3G3S1 (21.0%) Supplementary Fig. S2 and Supplementary Table S2). In terms of sialylation, 98.7% o the identified glycans were sialylated, with 51% bing mono-sialylated, 41.3% d-sialylated and 6.4% ti-sialylated (Table 2). In contrast, the rAAT before modification only had 47.9% total sialylation content with the major N-glycans being FA2G2S1 (18.9%),FA2G2 (17.1%) nd FA2G2S2 (10.5%) Table 2 and Supplementary Table S3); 26.6% wre mono-sialylated, 16.9% wre di-sialylated and 4.4% tri-sialylated (Table 2). Overall, we observed an approximately two-fold increase in the sialylation content of the modified rAAT and have successfully demonstrated the applicability of CMBs to purify glycans from recombinant therapeutic proteins thereby having confidence in the method’s capability to also isolate N-glycans from other biotherapeutic modalities that have complex glycan profiles.
CMBs enables automated high throughput purification and profiling of released N-glycans via a robotic liquid handler system
For the generation of an automated high throughput purification and profiling method for N-glycans, we adapted our workflow, including the release, labelling, and CMBs cleanup of the N-glycans, onto the robotic liquid handler platform. We performed 8 repetitions of the run using N-glycans released from hsIgG, tagged with RFMS and evaluated the performance of the automation against manual sample preparation. We found high reproducibility in using automation, with an average of 0.85% RSD, compared to manual procedures of 2.54% (n = 4) (Fig. 4). Furthermore, automation did not affect the sensitivity of the technique, as we found that both automated and manual procedures yielded the same distribution of glycans identified (Fig. 4). This suggests the potential capability of incorporating our CMB-based workflow in high-throughput glycomic approaches, where such fast and robust procedures are needed in the characterization of biotherapeutic products.
Comparison of the CMBs purification of hsIgG performed either manually or automated on the robotic liquid handler. Relative abundances of individual glycans purified using manual (green) or automated (yellow) procedures were quantified and compared, together with their RSDs. Error bars represent mean ± SD (n = 4 for manual preparation and n = 8 for automated preparation).
Discussion
Cellulose, in the form of suspension or packed into a filter tip for the purification and enrichment of glycan and glycopeptides has previously been reported27,28,43. However, the methods can be laborious and time consuming due to packing of the cellulose into the tips and could present inconsistency issues when handled and packed by different laboratory personnel. Cellulose, in the form of functionalized magnetic beads, on the other hand, help to mitigate the reproducibility issues and provides the benefit of high throughput analysis across multiple sample types, of both sample volume and fluorescent label, giving the flexibility for scaling down or scaling up. However, the disadvantage of using CMBs is that time and mixing is required for the binding and elution of glycans from the beads. We believe this can possibly be overcome by the reduction of the size of the magnetic beads thereby increasing the surface area of contact of the beads with the sample or increasing the amount of beads used. Using more beads could lead to an increase in the cost of purification but the overall cost would still remain low compared to usage of commercial purification cartridges or plates. We performed an assessment of the cost effectiveness of the CMBs and as a basis of comparison, we compared these costs across the three purification products purchased commercially, assuming a similar profit margin across them. The cost of purification per sample was Singapore dollar (SGD) 20.0 for the HILIC-µSPE plate, SGD 12.6 for G-10 column and lastly, SGD 1.4 for CMBs (per vial of CMBs can purify up to 90 samples based on 200 µg of beads per sample). Therefore, the CMBs workflow would lower the cost of glycosylation particularly in high-throughput applications.
The purification of glycans after their release from the protein and labelling is an essential step in glycosylation analysis as it eliminates background interferences and enhances their signal sensitivity and detection, especially for glycans present in low abundances that could trigger immunogenicity in humans such as N-glycans with galactose-alpha-1,3-galactose (alpha-gal)44,45 and N-glycolylneuraminic acid (Neu5Gc)45,46. Commercially available purification cartridges and plates are either expensive, limited by sample volume and/or do not cater for high throughput analysis. Here we capitalized on the highly hydrophilic properties of cellulose functionalized magnetic beads to bind to polar molecules and develop a cost-effective and automated high throughput workflow to purify fluorescent labelled N-glycans from biotherapeutic modalities. The combination of cellulose and magnetic nanoparticles presents an attractive alternative to current purification strategies as cellulose is readily available, affordable, biodegradable and sustainable in long term. The magnetic nanoparticles with their superparamagnetic properties, on the other hand, can be utilized easily in magnetic separation procedures to purify and isolate the molecules of interest. As such, they can be easily adapted for automation since no centrifugation or vacuum filtration is required, simplifying the whole purification process and making the approach scalable to cater to large volume of samples.
We have demonstrated that the purification strategy is equivalent to current techniques, and also displayed high compatibility with different fluorescent labels with good reproducibility. The workflow can be automated onto the liquid handler, allowing high throughput purification and profiling of N-glycans for biotherapeutic modalities. With the affordability of CMBs, the technique provides a cost-effective and environment-friendly alternative, sustainable in the long run. We believe with the versatility of our approach, there could be other extended applications such as its use to glycopeptides enrichment. Given its efficiency, we believe our approach described here provides a powerful supplement to the arsenal of techniques used in glycomic characterisation.
Materials and methods
Release of N-glycans and fluorescent labelling
N-glycans were released from human serum IgG and bovine fetuin (15 µg, both from Sigma-Aldrich, St Louis, MO, USA, cat. number I4506 and F3004). The glycoproteins were dried down and reconstituted in water (22.8 µL) and denatured using Rapigest™ SF (5% w/v, 6 µL) (Waters Corporation, Milford, MA, USA, cat. number 186001861) at 90 ºC for 10 min. After the denatured proteins have cooled to room temperature, PNGase F (600 U, 1.2 µL) (New England Biolabs, Ipswich, MA, USA, cat. number P0709L) was added with incubation at 55 ºC for 10 min to release the N-glycans. RFMS label (Waters Corporation, Milford, MA, USA, cat. number 186008091-1) was reconstituted in 131 µL anhydrous dimethyl formamide (DMF) and 12 µL of the label was added to the N-glycans followed by incubation at room temperature for 10 min.
For labelling with ProC (Sigma-Aldrich, St Louis, MO, USA, cat. number SML2088) and 2-AB (Acro Organics, Geel, Belgium, cat. number 104901000), the labelling protocol was as previously described16. In brief, the labelling mixture was prepared freshly by either dissolving ProC (38.3 mg/mL) or 2-AB (19.2 mg/mL) and 2-picoline borane (44.8 mg/mL) in dimethylsulfoxide (DMSO) and glacial acetic acid (70 : 30, v/v). The label (25 µL) was then added to the released N-glycans and incubated at 65 ºC for 2 h.
Purification of fluorescent derivatized N-glycans using hydrophilic 96-well micro SPE
After fluorescent derivatization, RFMS labelled N-glycans were diluted with 358 µL acetonitrile (ACN) and purified with Glycoworks HILIC µElution plate (Waters Corporation, Milford, MA, USA, cat. number 186002780) according to the manufacturer’s protocol (Waters Corporation). Each well was equilibrated with water (200 µL), followed by 85% (v/v) ACN (200 µL). The N-glycans were loaded to the well and the well was washed twice with 90% (v/v) ACN containing 1% formic acid (600 µL). Elution of the N-glycans was accomplished by addition of 3 × 30 µL of the GlycoWorks SPE elution buffer (Waters Corporation, Milford, MA, USA, cat. number 186007992). The eluted glycans were subsequently dried down in a vacuum centrifuge and reconstituted in a buffer consisting of water, dimethylformamide (DMF) and ACN (in ratio of 9 : 10 : 21, v/v) (40 µL) for analysis.
Purification of fluorescent derivatized N-glycans using PD MiniTrap G10 column
PD MiniTrap G10 column (Cytiva, Marlborough, MA, USA, cat. number GE28-9180-10) was equilibrated with 8 mL of water under gravity flow. RFMS labelled N-glycans were diluted with water (final volume of 300 µL) and loaded onto the column. The column was rinsed with 400 µL of water and N-glycans were eluted from the column with 600 µL of water. The eluted glycans were subsequently dried down and reconstituted as described in the HILIC-SPE procedure.
Purification of fluorescent derivatized N-glycans using cellulose magnetic beads (CMBs)
Fluorescent derivatized N-glycans were diluted with ACN to a final concentration of 90% (v/v) ACN. Storage buffer from the CMBs (200 µg) (Life Technologies Corporation, Carlsbad, CA, USA, cat. number, 4489112) was first removed and the beads were washed twice with water before being added to the labelled N-glycans, incubated at 24 °C with mixing on the Thermomixer® C (Eppendorf, Hamburg, Germany) at 1200 rpm for 5 min. After 5 min, the beads were immobilized onto a magnetic stand and the supernatant was removed. The beads were rinsed twice with 200 µL of 90% (v/v) ACN. N-glycans were eluted from the beads by addition of 200 µL of water with incubation at 24 °C and mixing at 1200 rpm for 5 min. The beads were immobilized and the purified N-glycans were collected and dried down using a vacuum centrifuge. 2-AB and ProC labelled N-glycans were reconstituted in 40 µL of 75% (v/v) ACN while RFMS labelled N-glycans were reconstituted as described in the HILIC-SPE procedure for injection into the LC-FLR-MS.
Automation of release of N-glycans, fluorescent labelling and purification procedure on liquid handler
The steps for release of N-glycans, RFMS labelling and their purification using CMBs were automated onto a robotic liquid handler (Tecan Freedom Evo 150, Tecan, Männedorf, Switzerland). The release of N-glycans from protein and RFMS labelling on the liquid handler was adapted from Waters Corporation47. In brief, the samples were dried down in a 96-well non-treated PCR plate. Water (10 µL) and Rapigest™ SF (3%w/v, 10 µL) were added to the samples in the plate and the plate was incubated at 90 ºC for 10 min for denaturation followed by addition of PNGase F (600 U, 10 µL) and incubation at 55 °C for another 10 min. After the plate is cooled for 5 min, RFMS label (10 µL), reconstituted in 110 µL anhydrous DMF, was then added and incubated at room temperature for at least 10 min.
CMBs (200 µg) were aliquoted into a 2 mL 96 deep-well non-treated plate. The storage buffer was removed and the beads washed twice and the RFMS labelled samples were transferred from the PCR plate to the deep-well plate containing the CMBs. ACN was added to the plate to a final concentration of 90% (v/v). The plate was then transferred manually to an external mixer for mixing on the Thermomixer® C at 1200 rpm for 5 min due to the speed limitation of the mixer on the workstation. After 5 min, the plate was transferred back to the liquid handler where subsequent removal of the supernatant and the washing steps were carried out. For elution, the mixing was similarly carried out on the external mixer before being transferred back to the workstation for collection of the eluate onto a new plate.
LC-FLR-MS analysis of N-glycans
Fluorescent derivatized N-glycans were analysed on a H-class Acquity UPLC system (Waters Corporation) consisting of a quaternary solvent manager, sampler manager and fluorescence detector coupled to a Xevo G2-S Q-TOF MS (Waters Corporation) system.
The labelled N-glycans were separated on a Waters Acquity UPLC glycan BEH amide column (2.1 mm x 150 mm, 1.7 μm) with mobile phase A comprising of 50 mM ammonium formate at pH 4.4 and ACN as mobile phase B. The separation gradient was 75 − 51% B at a flow rate of 0.4 mL/min for 40 min with the temperature of the column maintained at 60 °C. N-glycans were detected on the fluorescence detector at an excitation wavelength of 265 nm and emission wavelength of 425 nm for RF-MS labelled glycans, at 310 nm and 370 nm for ProC labelled glycans, and at 250 nm and 428 nm for 2-AB labelled glycans. An external standard of RF-MS, 2-AB (both from Waters Corporation, Milford, MA, USA, cat. number 186006841, 186007982) and ProC (Ludger, Oxfordshire, UK, cat. number CPROC-GHP-30) labelled dextran ladder was used respectively to establish a calibration curve for the different fluorescently labelled N-glycans from which the retention time of each chromatographic peak was converted into glucose units (GU).
Full MS scan data was acquired in positive ion mode in the range of 100 to 2,000 m/z using a capillary voltage of 2.75 kV, with the cone voltage set to 15 V, source temperature at 120 °C, desolvation gas flow at 800 L/H, desolvation temperature at 300 ˚C and scan time of 1 s. Glu-fibrinopeptide (m/z 785.8421) was used as a lock spray mass to calibrate the mass accuracy of the instrument during the analytical runs.
Data processing and structural assignment of N-glycans
Data acquisition and processing was performed using UNIFI scientific information system software (version 1.8.2.169, Waters Corporation). The retention time of each chromatographic peak was processed and converted into GU values and structural assignment of each peak was performed by matching their GU values against commercial library databases. For GU values that correspond to multiple structures, the MS data was used in addition to assign the correct structure based on mass confirmation (mass accuracy of 5 ppm). Relative abundances of each glycan was calculated and normalized by dividing the area of each glycan peak over the total area of all the glycans identified in the chromatograph.
Data availability
The data presented in this study are available on request from the corresponding author.
References
Varki, A. Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, 2022).
Boune, S., Hu, P., Epstein, A. L. & Khawli, L. A. Principles of N-linked glycosylation variations of igg‐based therapeutics: Pharmacokinetic and functional considerations. Antibodies 9, 1–20. https://doi.org/10.3390/antib9020022 (2020).
Falck, D. et al. Glycoform-resolved pharmacokinetic studies in a rat model employing glycoengineered variants of a therapeutic monoclonal antibody. MAbs 13, 145 (2021).
Luo, C. et al. Glycoengineering of pertuzumab and its impact on the pharmacokinetic/pharmacodynamic properties. Sci. Rep. 7, 859 (2017).
Chia, S. et al. Enhancing pharmacokinetic and pharmacodynamic properties of recombinant therapeutic proteins by manipulation of sialic acid content. Biomed. Pharmacother. 163, 56. https://doi.org/10.1016/j.biopha.2023.114757 (2023).
Liu, L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J. Pharmaceut. Sci. 104, 1866–1884. https://doi.org/10.1002/jps.24444 (2015).
Yamane-Ohnuki, N. & Satoh, M. Production of therapeutic antibodies with controlled fucosylation. mAbs 1, 230–236. https://doi.org/10.4161/mabs.1.3.8328 (2009).
Golay, J., Andrea, A. E. & Cattaneo, I. Role of Fc core fucosylation in the effector function of IgG1 antibodies. Front. Immunol. 13, 56. https://doi.org/10.3389/fimmu.2022.929895 (2022).
Mössner, E. et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell—mediated B-cell cytotoxicity. Blood 115, 4393–4402 (2010).
Román-Carrasco, P. et al. The α-Gal Syndrome and Potential Mechanisms. Front. Allergy 2021, 2. https://doi.org/10.3389/falgy.2021.783279 (2021).
Mimura, Y. et al. Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy. Protein Cell 9, 47–62. https://doi.org/10.1007/s13238-017-0433-3 (2018).
Pabst, M. et al. Comparison of fluorescent labels for oligosaccharides and introduction of a new postlabeling purification method. Anal. Biochem. 384, 263–273 (2009).
Doherty, M. et al. Plasma N-glycans in colorectal cancer risk. Sci. Rep. 8, 45 (2018).
Adamczyk, B., Stöckmann, H., O’Flaherty, R., Karlsson, N. G. & Rudd, P. M. High-throughput analysis of the plasma N-glycome by UHPLC. In Methods in Molecular Biology vol. 1503 97–108 (Humana Press Inc., 2017).
Kozak, R. P., Tortosa, C. B., Fernandes, D. L. & Spencer, D. I. R. Comparison of procainamide and 2-aminobenzamide labeling for profiling and identification of glycans liquid chromatography with fluorescence detection coupled to electrospray ionization-mass spectrometry. Anal. Biochem. 486, 38–40 (2015).
Keser, T., Pavic, T., Lauc, G. & Gornik, O. Comparison of 2-aminobenzamide, procainamide and RapiFluor-MS as derivatizing agents for high-throughput HILIC-UPLC-FLR-MS N-glycan analysis. Front. Chem. 6, (2018).
Zhang, P. et al. Characterization of a GDP-fucose transporter and a fucosyltransferase involved in the fucosylation of glycoproteins in the diatom Phaeodactylum tricornutum. Front. Plant. Sci. 10, (2019).
Goh, J. S. Y. et al. Highly sialylated recombinant human erythropoietin production in large-scale perfusion bioreactor utilizing CHO-gmt4 (JW152) with restored GnT I function. Biotechnol. J. 9, 100–109 (2014).
North, S. J. et al. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285, 5759–5775 (2010).
Váradi, C., Lew, C. & Guttman, A. Rapid magnetic bead based sample preparation for automated and high throughput N-glycan analysis of therapeutic antibodies. Anal. Chem. 86, 5682–5687 (2014).
Szigeti, M., Guttman, A. & High-Throughput, N-G. Analysis with Rapid Magnetic Bead-Based Sample Preparation 265–272 (2017). https://doi.org/10.1007/978-1-4939-6493-2_19.
Chen, X. & Flynn, G. C. Analysis of N-glycans from recombinant immunoglobulin G by on-line reversed-phase high-performance liquid chromatography/mass spectrometry. Anal. Biochem. 370, 147–161 (2007).
Yang, X. et al. Ultrafast and high-throughput N-glycan analysis for monoclonal antibodies. MAbs 8, 706–717 (2016).
Khatri, K., Klein, J. A. & Zaia, J. Use of an informed search space maximizes confidence of site-specific assignment of glycoprotein glycosylation. Anal. Bioanal Chem. 409, 607–618 (2017).
Tu, H. et al. A reference standard for analytical testing of erythropoietin. Pharm. Res. 39, 553–562 (2022).
Verostek, M. F., Lubowski, C. & Trimble, R. B. Selective Organic Precipitation/Extraction of released N-Glycans following large-scale enzymatic deglycosylation of glycoproteins. Anal. Biochem. 278, 111–122 (2000).
Ruhaak, L. R. et al. Hydrophilic interaction chromatography-based high-throughput sample preparation method for N-glycan analysis from total human plasma glycoproteins. Anal. Chem. 80, 6119–6126 (2008).
Selman, M. H. J., Hemayatkar, M., Deelder, A. M. & Wuhrer, M. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83, 2492–2499 (2011).
Wuhrer, M., De Boer, A. R. & Deelder, A. M. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass Spectrom. Rev. 28, 192–206 (2009).
Lauber, M. A. et al. Rapid preparation of released N -glycans for HILIC analysis using a labeling reagent that facilitates sensitive fluorescence and ESI-MS detection. Anal. Chem. 87, 5401–5409 (2015).
Stöckmann, H., Adamczyk, B., Hayes, J. & Rudd, P. M. Automated, high-throughput IgG-antibody glycoprofiling platform. Anal. Chem. 85, 8841–8849 (2013).
Váradi, C., Sikora, E., Vanyorek, L. & Viskolcz, B. Purification of fluorescently derivatized n-glycans by magnetic iron nanoparticles. Nanomaterials 9, (2019).
Shubhakar, A. et al. High-throughput analysis and automation for Glycomics studies. Chromatographia 78, 321–333 (2015).
Molares Vila, A., de Rupérez Pérez, P. & Caso Peláez, E. Gago-Martínez, A. Development of a new magnetic beads-based immunoprecipitation strategy for proteomics analysis. J. Proteom. 73, 1491–1501 (2010).
Socas-Rodríguez, B., Herrera-Herrera, A. V., Asensio-Ramos, M. & Rodríguez-Delgado, M. Á. Recent applications of magnetic nanoparticles in food analysis. Processes 8, 1140 (2020).
Tang, C. et al. Application of magnetic nanoparticles in nucleic acid detection. J. Nanobiotechnol. 18, 62 (2020).
Materón, E. M. et al. Magnetic nanoparticles in biomedical applications: a review. Appl. Surf. Sci. Adv. 6, 100163 (2021).
Mariño, K., Bones, J., Kattla, J. J. & Rudd, P. M. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6, 713–723. https://doi.org/10.1038/nchembio.437 (2010).
Ltd, L. Fetuin N-Glycan Library Table. www.ludger.com (2022).
Ltd, L. LudgerTag Procainamide Glycan Labelling Kit (2022).
Kim, A. et al. Peptide-N-glycosidase F or A treatment and procainamide-labeling for identification and quantification of N-glycans in two types of mammalian glycoproteins using UPLC and LC-MS/MS. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 1214, 145 (2023).
Pallister, E. G. et al. Exploiting the disialyl galactose activity of α2,6-sialyltransferase from photobacterium damselae to generate a highly sialylated recombinant α-1-antitrypsin. Biochemistry 59, 3123–3128 (2020).
Sha, Q. et al. Cellulose microspheres-filled pipet tips for purification and enrichment of glycans and glycopeptides. J. Chromatogr. A 1569, 8–16 (2018).
Chinuki, Y. & Morita, E. Alpha-gal-containing biologics and anaphylaxis. Allergology Int. 68, 296–300 (2019).
Yu, C. et al. At least two fc Neu5Gc residues of monoclonal antibodies are required for binding to anti-Neu5Gc antibody. Sci. Rep. 6, 20029 (2016).
Ghaderi, D., Taylor, R. E., Padler-Karavani, V., Diaz, S. & Varki, A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat. Biotechnol. 28, 863–867 (2010).
Ds, W. O. R., Koza, S. M., Mccall, S. A., Lauber, M. A. & Chambers, E. E. Quality Control and Automation Friendly GlycoWorks RapiFluor-MS N-Glycan Sample Preparation (2022).
Acknowledgements
We would like to acknowledge and thank Terry Nguyen-Khuong for his contributions and advice in the initial development of this study.
Funding
This work is funded and supported by Agency for Science Technology and Research (A*STAR), under the grant C210112057 awarded to I.W.
Author information
Authors and Affiliations
Contributions
Conceptualization, C.W; methodology, C.W. and S.C.; validation, formal analysis, investigation, S.J.T and C.W.; resources, T.S.J., Q.B.Y. and C.W.; data curation, C.W.; writing – original draft preparation C.W. and S.C.; review and editing, S.C., I.W. and C.W.; visualization, T.S.J. and C.W.; supervision, I.W and S.C. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wan, C., Tay, S.J., Qiu, B. et al. Cellulose functionalized magnetic beads for high throughput glycosylation analysis in biotherapeutic modalities. Sci Rep 14, 29735 (2024). https://doi.org/10.1038/s41598-024-80649-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80649-y