Abstract
The application of pesticides may adversely impact a variety of non-target organisms. The use of side-effect-free herbal remedies to protect against the toxicity of harmful pesticides such as etoxazole has gained attention in recent times. The current study aimed to reveal the potential mitigating efficacy of Achillea millefolium L. extract against etoxazole toxicity in Allium cepa L. A. cepa bulbs in the control group were applied with tap water, while bulbs in the treatment groups were applied with etoxazole at dose of 0.5 m/L and two different doses of A. millefolium extract (200 mg/L and 400 mg/L). The impact of the treatments on certain parameters was evaluated. The molecular docking analysis was employed to investigate the potential interactions of etoxazole with DNA species, DNA topoisomerases, tubulin proteins, glutamate-1-semialdehyde aminotransferase, and protochlorophyllide reductase. The phenolic profile of A. millefolium was assessed. Etoxazole exposure reduced rooting percentage, root length, weight gain, mitotic index, and levels of chlorophyll a and chlorophyll b. Conversely, etoxazole treatment led to an increase in chromosomal aberrations and micronuclei occurrence. The most frequently observed chromosomal aberrations induced by etoxazole, which serve as bioindicators of genotoxicity, were fragment, vagrant chromosome, sticky chromosome, unequal chromatin distribution, bridge, reverse polarization, and vacuolated nucleus. The levels of malondialdehyde and antioxidant enzyme (superoxide dismutase and catalase) activities were also elevated. Epidermis cell damage, flattened cell nucleus, thickened cortex cell wall, and thickened conduction tissue were the meristematic cell disorders triggered by etoxazole. Molecular docking studies showed that etoxazole can interact directly with DNA, tubulins, and the enzymes mentioned above. A. millefolium extract was found to contain a substantial quantity of phenolic compounds. A. millefolium extract, when co-administered with etoxazole, attenuated all toxic effects of etoxazole dose-dependently. In conclusion, A. millefolium may potentially serve as a reliable pharmacological shield against the toxicity of pesticides in non-target organisms.
Similar content being viewed by others
Introduction
Over the past few decades, pesticides have been widely employed in the agricultural sector to avoid yield loss. Many organisms that are not intended targets of pesticides suffer due to their widespread release into the environment. Since pesticides can be transported by air, water, spray drift, and post-application evaporation far from the application site, they can damage geographically remote habitats1. Therefore, a wide range of organisms, including plants, fishes, pets, beneficial insects, and humans, can be affected by the indiscriminate use of these chemicals. Human exposure to pesticides may occur through occupation or the food chain, and acute or chronic toxicity may develop over time2.
Etoxazole, also known as 2,4-diphenyl-1,3-oxazolines, is an efficient systemic acaricide to suppress the eggs, larvae, and nymphs of spider mites, including Tetranychus urticae and Panonychus ulmi3. As a narrow-spectrum organofluorine, it has been employed since 1998 to enhance the quality and yield of crops such as apples, citrus, pome fruits and cotton. Its mechanism of action involves the inhibition of chitin synthesis, which is required for exoskeleton formation4. The inhibition process is achieved through the binding of etoxazole specifically to the sulfonylurea receptor5. Due to its half-life of about 20 days and high bioaccumulation tendency, etoxazole represents a significant risk even to individuals who do not have direct contact with the chemical4. In previous studies, etoxazole was reported to be neurotoxic6, cytotoxic7and genotoxic8in non-target organisms. Furthermore, the available literature indicates that it may impair cardiovascular health9and induce oxidative stress10.
The risks to human health are growing due to industrialization and technological advancements. Synthetic drugs, which often lead to undesirable consequences, are less preferred in the modern era. Despite significant breakthroughs in recent pharmaceutical technology, 60% of the 300,000 seed plants on earth are used for medicinal purposes11. Indeed, various parts of medicinal plants, which contain a plethora of many bioactive compounds, have been recognized as stand-alone treatments and in combination with medical applications12. Achillea millefolium L. (yarrow), a member of the Asteraceaefamily, is a perennial herb that has been used medicinally for centuries13. A. millefoliumhas a long history of use in the treatment of spasmodic gastrointestinal disorders, gynecological disorders, hepatobiliary, wound treatment, malaria, hepatitis, and jaundice14. Its antioxidant, antibacterial, anti-inflammatory, anti-hepatotoxic, and anticancer activities are attributed to the high amount of polyphenolic and flavonoid compounds it contains15. The results of animal experimentation have demonstrated that this plant is generally safe16. Additionally, reports have indicated that the antioxidant compounds present in A. millefoliumexert an anti-genotoxic effect17,18. The bioactive content of A. millefoliumalso functions as singlet oxygen quenchers, reductants, and hydrogen donors against reactive molecules that induce oxidative stress and ultimately cause cell death15.
Allium cepa L., one of the organisms that helps to determine cytotoxicity and genotoxicity caused by chemicals and complex mixtures, is a valuable model for assessing in vivo damage. The Allium assay, initially developed by Levan in 1938 to monitor mitotic spindle disorders in Alliumroot cells exposed to colchicine, is a highly effective, easy, cheap, and short-term system that provides data compatible with mammalian test systems19. The success of the assay, which demonstrated up to 82% agreement with test systems involving rodents, can be attributed to the fact that A. cepais a multicellular, and eukaryotic organism. This test, which is used to determine the impact of environmental mutagens on the mitotic phase of root meristem cells, also allows the observation of chromosomal abnormalities and micronucleus formation20.
Etoxazole pesticide accumulation or contamination into the environment endangers several non-target species. It is crucial to assess the efficacy of commonly utilized pesticides in living systems, as this enables an accurate determination of the extent of damage to plants and other biological organisms. This process also serves to reinforce the prudent deployment of these pesticides in agricultural practices. The literature provides rich information on the antioxidant and antigenotoxic effects of A. millefolium. However, the protective mechanism of the plant against pesticide-induced metabolic disorders has not been fully elucidated. Therefore, it is crucial to diversify research in these areas. In the present study, a multifaceted approach was employed to investigate the potential alleviatory effect of A. millefolium extract against the toxicity caused by etoxazole in A. cepa, a well-established model organism for assessing the toxic effects of hazardous pesticides. In order to achieve this, a series of analyses were conducted on A. cepa roots exposed to etoxazole and A. millefolium extract, encompassing growth physiology, biochemical, genotoxicity, and meristematic cell damage assessments. The phenolic compounds contained in the extract were identified to clarify the protective mechanism of A. millefolium. Furthermore, possible molecular mechanisms of etoxazole toxicity were investigated through molecular docking analysis.
Materials and methods
Preparation of experimental design, materials, and test chemicals
A. cepa bulbs, carefully selected for their close initial weights (10.6–12.0 g), were obtained from a local market in Giresun, Türkiye. The bulbs were stripped of their outer leaves and old roots in the laboratory, and six groups were formed with these bulbs. Throughout the experiment, the groups were exposed to tap water (the control group), 200 mg/L A. millefolium extract (the AME 1 group), 400 mg/L A. millefolium extract (the AME 2 group), 0.5 mL/L etoxazole (the ETX group), 0.5 mL/L etoxazole + 200 mg/L A. millefolium extract (the ETXAME 1 group) and 0.5 mL/L etoxazole + 400 mg/L A. millefolium extract (the ETXAME 2 group), respectively (Table 1). Onion bulbs were placed in tubes with the disc stem parts in contact with the solutions. Extraction was performed in a shaker incubator for 2 h at room temperature by adding 10 g of plant material to 100 mL of water. Following incubation, the solid particles were separated by filtration, and the resulting liquid was centrifuged at 10,000 rpm for 10 min. The supernatant was evaporated and the pellet was collected for further examination, ensuring the highest standards of preparation and reliability in our experiment.
The study of Macar et al.8 was considered in dose selection for etoxazole. For the preparation of the aqueous solution, an etoxazole solution (manufactured by Hektaş Ticaret Türk A.Ş., Kocaeli, Türkiye and trade name Delos) with 110 g/L of active ingredient was used. When determining the appropriate doses of A. millefoliumextract, the dosages used in the limited number of studies documented in the literature and shown to have a protective effect were taken into account21. Upper leaves and inflorescences of A. millefolium were gathered from Çambaşı / Ordu and identified in the Biology Laboratory of Giresun University. After washing and drying, the plant material was mixed and homogenized. Identification of A. millefolium was made in the Department of Botany of Giresun University (Gaziler District, Prof. Ahmet Taner Kışlalı Avenue, postal code: 28200 Giresun/Türkiye) and a sample was archived in the herbarium.
Test solution applications continued for 72 h to collect the root tissues needed for the analyses. The duration of application was prolonged to 144 h to obtain leaf tissues for chlorophyll analysis. The tubes containing the bulbs were left in the dark and at room temperature while they underwent chemical treatment. The solutions in contact with the bulbs were changed daily to prevent concentration variations.
Methods for analyses of physiological parameters
Bulbs having adventitious roots of at least 1 cm were deemed “rooted” to calculate (Eq. 1) the rooting percentage22.
Weight increase was determined by calculating the difference (g) between the bulbs’ initial weight prior to chemical treatment and their final weight (g) after the treatment (72 h). A precision scale was employed for weighing.
After 72 h of chemical treatment, the root length (cm) was measured by placing a ruler between the root tip and the root base.
Methods for analyses of genotoxicity parameters
Mitotic index (MI), micronucleus (MN) frequency, and number of chromosomal aberrations (CAs) in each group were examined in order to comprehend etoxazole-induced genotoxicity and the impact of A. millefolium against it. As recommended by Staykova et al.23, Clarke’s fixator (glacial acetic acid (V) + ethyl alcohol (3 V) was utilized to fix root tips (1.5 cm) to assess MI, MN, and CAs scores. The fixation solution was removed by keeping the roots in 96% ethyl alcohol for 10 min. Roots were then placed in 1 N HCl solution at 62 °C for 14 min. Subsequently, the root tips were transferred to petri dishes filled with 45% glacial acetic acid and left for 30 min. The root tips were soaked in acetocarmine for 12 h, and then crushed between a coverslip and a slide to finish the preparations. MN and CA abundance and mitotic cells on the squash preparations were examined using the Irmeco IM-450 TI type microscope (500X magnification). The regions selected for analysis on the slides were chosen at random. MN and CAs were calculated by analyzing 100 cells per slide prepared from a single root tip and 1,000 cells from 10 root tips per group. Equation 2 reflects the MI (%) computation. MI was calculated by analyzing 1,000 cells per slide prepared from a single root tip and 10,000 cells from 10 root tips per group.
The MN determination was conducted following the guidelines set out by Fenech et al.24. Extra care was taken to distinguish the MN from the nuclear bud.
Method for molecular docking analysis
Potential interactions between etoxazole and DNA molecules, DNA topoisomerases, tubulins, glutamate-1-semialdehyde aminotransferase, and protochlorophyllide reductase were examined using molecular docking. The protein data bank provided the following 3D structures: B-DNA dodecamer (PDB ID: 1BNA)25, B-DNA dodecamer d (PDB ID: 195D)26, DNA (PDB ID: 1CP8)27, DNA topoisomerase I (PDB ID:1K4T), DNA topoisomerase II (PDB ID:5GWK)28,29, tubulin proteins (alpha-1B chain and tubulin beta chain) (PDB ID: 6RZB)30, glutamate-1-semialdehyde aminotransferase (PDB ID:2ZSL)31and protochlorophyllide reductase (PDB ID:6R48)32. The 3D configuration of etoxazole with 153,974 PubChem CID was obtained from PubChem. Firstly, the active sites of the protein molecules were recognized. Water molecules and ligands were then removed from their structure, and polar hydrogen atoms were added. Thus, the proteins became ready for the molecular docking process. Gromos 43B1 with Swiss-PdbViewer software (v.4.1.0)33was used to reduce the energy of the proteins, and Open Babel software (v.2.4.0)34was utilized to decrease the energy of the uff-force field in the 3D structure of etoxazole molecule. Etoxazole was allocated Gasteiger charges, while receptor molecules were assigned to Kollman charges. A grid box including all structures of DNA and the active sites of proteins was used in the molecular docking process. Docking was carried out using the Autodock 4.2.6 program using the Lamarckian genetic algorithm35. The Biovia Discovery Studio 2020 Client was employed to accomplish post-docking arrangements and 3D visualizations36 (Biovia, 2023).
Methods for analyses of biochemical parameters
Chlorophyll a and chlorophyll b contents of fresh leaves were assessed following the 144-hour treatment period. Pigments were isolated and determined based on the method described by Kaydan et al.37. 2.5 mL of 80% acetone dissolved the pigments in 0.1 g leaf samples. The leaves were first ground precisely in the solution and then allowed to wait for seven days in a refrigerator. The waiting procedure took place in complete darkness. After filter paper was used to remove any remaining solid particles, 2.5 mL of 80% acetone was added to the extract tubes. The extracts were centrifuged at 3000 rpm without allowing the acetone to evaporate. The supernatant was collected, and the procedure was repeated after adding 2.5 mL of 80% acetone to the supernatant. Using a UV-Vis spectrophotometer set to wavelengths of 663 nm and 645 nm, the absorbance of the supernatant was measured38.
OD 645 and OD 663 are the sample absorbance measurements at 645 and 663 wavelengths, respectively. W is the weight of the fresh leaf sample (g); V is the total volume (mL) of 80% acetone with the sample.
The method suggested by Bates et al.39 was employed to assess the proline contents in the groups. Following the completion of the treatments, 10 mL of 3% aqueous sulfosalicylic acid was added to 0.5 g of root samples to homogenize the roots using a mortar and pestle. Whatman No. 2 filtering was used to get the filtrate from this homogenate. The filtrate (2 mL) was mixed with the same volume of glacial acetic acid (2 mL) and an acid-ninhydrin combination (2 mL). The tubes containing the new mixture were kept at 100 °C for 1 h before being totally cooled. After adding 4 mL of toluene, the tubes were stirred for 10–15 s. As a result, the toluene-dissolved chromophore fraction was separated from the remaining phase. Using a UV-Vis spectrophotometer set to a wavelength of 520 nm, the absorbance of the chromophore fraction was read. Using Eq. 5 and the free proline standard curve, proline contents were computed.
The damaging impact of etoxazole-induced oxidative stress on membranes and the ability of A. millefolliumto guard against it were investigated using malondialdehyde (MDA) assay40. Following a homogenization process for 0.5 g of root sample in 1 mL of trichloroacetic acid (TCA) (5%) solution, the supernatants were centrifuged at 12,000 rpm for 15 min to ascertain the amount of MDA. Then, the supernatant was mixed with the same volume of 0.5% thiobarbituric acid and 20% TCA, and the mixture was heated to a boiling temperature for 25 min. The reaction medium was centrifuged at 10,000 rpm for 5 min after the reaction was stopped by submerging the tubes in an ice bath. Using a UV-Vis spectrophotometer set to a wavelength of 532 nm, the absorbance of the supernatant was read to calculate the MDA concentration as µM/g FW.
Superoxide dismutase (SOD) and catalase (CAT) enzyme activity were two additional metrics utilized to assess etoxazole-induced oxidative stress. For the extraction of enzymes, 0.25 g of root material was collected. Then, using the protocol recommended by Zou et al.41, the root tissue was homogenized in 2.5 mL of ice-cold sodium phosphate buffer with pH 7.8. The supernatant, obtained by separating it with a 20-minute centrifugation at 10,000 rpm, was used to assess the enzyme activities.
The reaction medium for SOD activity was prepared by adding methionine, riboflavin, nitroblue tetrazolium chloride, polyvinylpyrrolidone, EDTA-Na2, and deionized water to a sodium phosphate buffer (pH 7.8) containing tube42. Two 15-watt fluorescent lights were used to illuminate the mixture for 10 min after 0.01 mL of supernatant was added. Following this procedure, the reaction mixture was kept in a dark chamber for 15 min to terminate the reaction. Using a UV-Vis spectrophotometer set to wavelength of a 560 nm, the absorbance of the mixture was read to calculate the SOD activity as U mg/FW.
The reaction medium for CAT activity was prepared by adding deionized water and hydrogen peroxide (H2O2) to a sodium phosphate buffer-containing tube43. To initiate the enzymatic reaction with H2O2, 0.2 mL of supernatant was mixed with the medium. As H2O2 dropped, absorbance at 240 nm reduced as well. The final unit for CAT activity was OD240 nm min/g FW.
Method for analysis of meristematic cell damages
The effects of etoxazole and A. millefoliumon root meristem cells were examined by randomly selecting ten germinated onions from each group8. Hand-taken cross-sections from randomly chosen adventitious roots were used to create ten slides in total for each group. Sections were stained on a slide with a 1% methylene blue stain. A research microscope at a magnification of ×400 was employed to screen and photograph the meristematic cells. The cells were classified as extremely damaged, moderately damaged, slightly damaged, and undamaged.
Phenolic composition analysis of A. millefolium extract
The antioxidant power of herbal extracts depends mainly on their phenolic composition. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to determine the quantitative phenolic composition of A. millefolium. HUBTUAM Laboratory (Hitit University) completed the assessment. The sample (1 g) was extracted in an ultrasonic bath for 2 h using methanol/dichloromethane (4 V : V) as a solvent and then filtered through a 0.45 m syringe filter. An ODS Hypersil column (4.6 × 250 mm) and a Thermo Scientific LC–MS/MS equipment (ICAP QC, USA) were utilized for the analysis. With a solvent flow rate of 0.7 mL/min and a column oven set to 30 °C, the analysis took 34 min to complete. The following are the criteria for the MS/MS analysis: The temperatures of the vaporizer and capillary are 350 °C and 300 °C, respectively. Sheat gas pressure and aux gas pressure are 30 Arb and 13 Arb, while the positive and negative polarities are 2500 V and 4 µA44,45.
Method for statistical analysis
The SPSS for Windows Ver. 22.0 package (SPSS Inc, USA, 2013) was utilized to analyze statistical data. Differences between groups were defined by performing one-way ANOVA followed by Duncan tests. The means of the data were deemed statistically significant if the p-value was less than 0.05 (p < 0.05).
Results and discussions
Etoxazole, A. millefolium and, a combination of etoxazole and A. millefolium treatments were tested for their effects on A. cepa root meristem cells by analyzing changes in specific physiological parameters (Table 2). A comparison of the AME 1 and AME 2 groups with the control group revealed that the doses of A. millefolium employed did not significantly affect the weight increase, root length, or rooting percentage. Thus, the extract did not induce any toxicity in A. cepa that would result in physiological damage. Conversely, all values exhibited a noteworthy reduction in the ETX group compared to the control group (p < 0.05). In the ETX group, the rooting percentage dropped to 65%. In this group, the root length and weight gain decreased by 69.4% and 64.8%, respectively, due to etoxazole treatment in comparison to the control. Growth retardation is regarded as one possible biochemical indicator of pesticide exposure46. A preceding study by Macar et al.8 also demonstrated that etoxazole impeded the growth of A. cepa. In the aforementioned study, the application of elevated pesticide doses served to exacerbate this effect. It has also been proven that etoxazole exerts a growth-suppressive effect on Danio rerio, a non-target organism, and this is achieved via inhibition of cell cycle-promoting genes and the induction of apoptosis9. Likewise, an increase in etoxazole concentration resulted in the observation of cytotoxicity in MCF-7 cells, which manifested as dose-dependent growth inhibition47. According to Park et al.4, many variables, such as cell cycle arrest, depolarization of the mitochondrial membrane potential, and activation of ER stress-response proteins, have been proposed as possible causes of the growth retardation observed in etoxazole-treated cells. Plant root growth is correlated with the degree of elongation that occurs during the differentiation step and the increase in cell quantity via mitosis. Indeed, pesticides may stunt plant growth by preventing the uptake of water and mineral components from the root environment, which is critical for cell division48. Anatomical defects in the cellular structure of etoxazole-treated roots may also lead to reduced growth directly or by interfering with water transport8. Pesticides can also retard root growth and suppress germination by interfering with the plant cell’s ability to produce energy and the synthesis of lipids, which are integral components of cell membranes49. When DNA stressors are present, reactive oxygen species (ROS) homeostasis mainly regulates root meristematic zone growth. It should also be noted that the growth of cells is halted in the presence of DNA damage, and a correlation has also been documented between the accumulation of ROS and DNA damage50. These results provide valuable insights into the effects of etoxazole and A. millefolium treatments on A. cepa root cells, contributing to understanding pesticide toxicity and potential mitigation strategies.
In the ETXAME 1 and ETXAME 2 groups, in which A. millefolium and etoxazole were administered as a combination, a significant enhancement in growth was observed in comparison to the ETX group (p < 0.05) (Table 2). Although the increases in the ETXAME 1 and ETXAME 2 groups were more pronounced as the dose of A. millefolium in the mixture increased, they remained below the levels observed in the control group. This is the first study to demonstrate the relieving function of A. millefolium against the growth-limiting effect of pesticide toxicity in A. cepa. This finding is particularly significant as it suggests a potential strategy for mitigating the growth-limiting effect of pesticide toxicity in A. cepa. Barut et al.51 reported that A. millefolium extract has a distinctive antioxidant composition and membrane-protective effect. Furthermore, it has been demonstrated that the bioactive constituents present in A. millefoliumextract can reduce DNA damage and influence the cell cycle and apoptosis17,52. It can be interpreted that these constituents may be linked to the restorative properties of A. millefolium and the growth retardation of A. cepa exposed to etoxazole.
Control (C): Tap water, AME 1: 200 mg/L A. millefolium extract, AME 2: 400 mg/L A. millefolium extract, ETX: 0.5 mL/L etoxazole, ETXAME 1: 0.5 mL/L etoxazole + 200 mg/L A. millefolium extract, ETXAME 2: 0.5 mL/L etoxazole + 400 mg/L A. millefolium extract. Data are expressed as mean ± SD. n = 50 for rooting percentage and n = 10 for root length and weight increase. Means with different letters (a−d) in the same column are significant at p < 0.05.
The effects of etoxazole, A. millefolium extract, and combinations of the two treatments on A. cepa root meristem cells were investigated by analyzing genotoxicity parameters, including MI, MN, and CAs (Table 3). The examination of the data from the AME 1, AME 2, and control groups revealed that exposure to A. millefolium extract did not induce any genotoxic effect on A. ceparoot cells. Conversely, the incidence of MI in the ETX group was 32% lower than that of the control group. MI is a metric that may be used to forecast the cytotoxicity of various chemicals and to quantify an organism’s mitotic activity. Apical meristematic cells undergo interphase and mitotic phases to finish the cell cycle, leading to roots forming. Therefore, MI and root growth rate can be employed in mutually reinforcing to quantify the extent of damage that a contaminant can inflict upon cell proliferation53. The findings of our study indicate that exposure to etoxazole leads to a simultaneous reduction in root growth and MI levels (Tables 2 and 3).
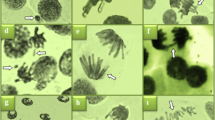
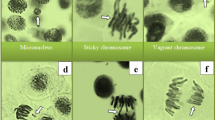
Notable elevations in MN and CAs levels were observed in the ETX group following the administration of etoxazole (Table 3). MN (Fig. 2a) is an abnormal nuclear body with an atypical nuclear envelope. It consists of chromosome fragments resulting from mitotic segregation failures or the absence of DNA repair. In A. cepameristematic cells, CAs and the development of MN have been employed as dependable biological markers and predictors of xenobiotic-related DNA damage53. Additionally, Khanna and Sharma54reported that a 45% decrease in the proportion of root development indicates that hazardous chemicals cause genetic alterations in plants. Indeed, growth suppression may result from CAs accumulation and a decline in mitotic activity53. The term CAs refers to a collection of differences in the quantity and/or morphology of chromosomes that are apparent during cell division55. In the ETX group, CAs induced by etoxazole treatment were sorted as fragment, vagrant chromosome, sticky chromosome, unequal chromatin distribution, bridge, reverse polarization, and vacuolated nucleus according to their abundance (Table 2). The most frequent chromosomal defect, fragment (Table 3; Fig. 2b), is brought on by either the disruption of double-stranded DNA or the suppression of DNA synthesis56. When failures in the spindle thread systems compel a chromosome to move toward the poles before the set of chromosomes of which it is a member, this chromosome is called a vagrant (Fig. 2c). In that case, an unbalanced chromatin distribution occurs in the daughter cells57. Indeed, the present study demonstrates that, in the etoxazole-treated group, unequal chromatin distribution (Fig. 2e), similar to vagrant chromosomes, was among the most prevalent CAs (Table 3). Another CA that increased in the etoxazole-treated group was the sticky chromosome (Table 3; Fig. 2d). Stickiness, a chromatid-type disorder, is an irreversible fatal error. It is attributed to several factors, including condensation and depolymerization of chromosomal DNA, inter-chromosomal entanglements, and chromosomes losing mobility58. It has been reported that ROS can interact with DNA molecules or histone proteins directly or indirectly, resulting in alterations to the surface properties of chromosomes and the induction of chromosome stickiness56. Additionally, ROS have been demonstrated to induce DNA strand breaks and irreparable damage to proteins engaged in DNA replication, recombination repair, and transcription, consequently leading to chromosomal aberrations59. According to Sabeen et al.58, sticky chromosomes have been demonstrated to result in anaphase failure, giving rise to the induction of chromosome bridging. Our findings showed that etoxazole is an inducer of bridge (Fig. 2f) formation in A. cepa root cells (Table 3). Barman and Ray60 suggested that the direct impact of a substance on the chromosome (DNA molecule) results in chromosomal fragments and bridges. Moreover, MN, bridge, and fragment are indicators that a chemical is clastogenic. Reverse polarization (Fig. 2g) was another type of chromosomal failure in our study. Kalcheva et al.61 stated that cells with reverse polarization and vagrant chromosomes are fundamentally caused by spindle system disruption. According to Seth et al.59, bridge and unequally distributed chromosomes are also among the spindle-related aberrations. In this case, etoxazole is a mitotic spindle poison for the root cells of A. cepa. In the ETX group, the vacuolated nucleus was the CAs with the lowest incidence (Table 3; Fig. 2h). Nuclear vacuolization is among the nuclear changes leading to apoptosis62, indicating that a nuclear toxin inhibits DNA production63. Our results agreed with those of Macar et al.8, who showed that MI decreased and MN and CAs accumulation increased in A. cepafollowing etoxazole treatment. Furthermore, many other pesticides have also been proven to reduce MI and/or induce the formation of MN and CAs in non-target organisms64,65. Park et al.9proved that etoxazole promotes ROS formation along with mitosis inhibition. In addition to direct interactions between pesticides and DNA, pesticide-induced oxidative stress is one of the most likely sources of genotoxicity. Extremely reactive radicals, including ROS, cause oxidative DNA lesions by interfering with cellular metabolism, compromising antioxidant defense, damaging macromolecules, including DNA, RNA, and protein, and impairing the activity of DNA repair proteins. Additionally, ROS have been shown to directly target double-stranded DNA and induce oxidative base modification, which can lead to DNA damage such as single- and double-stranded breaks66.
In the ETXAME 1 and ETXAME 2 groups, there was a notable reduction in the frequency of MN and CAs and a significant increase in the level of MI compared with the ETX group (p < 0.05) (Table 3). Although the values showing this alleviation did not reach the levels of the control group, it became more pronounced with increasing extract doses in the mixture of etoxazole and A. millefolium extract. Studies on the antigenotoxic potential of A. millefolium are extremely scarce in the literature. In a previous study, Shahani et al.17 demonstrated that A. millefolium extract possesses an antigenotoxic effect against ionizing radiation in human blood cells. Likewise, Montaser et al.67 indicated that an extract of A. millefolium effectively reduced the formation of MN and mutagenicity in human peripheral blood cell cultures. The researchers attributed these effects to the antioxidant properties of the extract.
Control (C): Tap water, AME 1: 200 mg/L A. millefolium extract, AME 2: 400 mg/L A. millefolium extract, ETX: 0.5 mL/L etoxazole, ETXAME 1: 0.5 mL/L etoxazole + 200 mg/L A. millefolium extract, ETXAME 2: 0.5 mL/L etoxazole + 400 mg/L A. millefolium extract. Data are expressed as mean ± SD. n = 10 for all parameters. Means with different letters (a−d) in the same row are significant at p < 0.05 level. MI: mitotic index, MN: micronucleus, FRG: fragment, VK: vagrant chromosome, SC: sticky chromosome, UDC: unequal chromatin distribution, B: bridge, RP: reverse polarization, VN: vacuolated nucleus.
Control (C): Tap water, AME 1: 200 mg/L A. millefolium extract, AME 2: 400 mg/L A. millefolium extract, ETX: 0.5 mL/L etoxazole, ETXAME 1: 0.5 mL/L etoxazole + 200 mg/L A. millefolium extract, ETXAME 2: 0.5 mL/L etoxazole + 400 mg/L A. millefolium extract. Data are expressed as mean ± SD. n = 10 for all parameters. Means with different letters (a−d) in the same column are significant at p < 0.05 level. MDA: Malondialdehyde, SOD: Superoxide dismutase, CAT: Catalase.
Molecular docking strategies are instruments for defining the complex relationships between tiny and macromolecules, providing important information about their possible impacts on biological processes. The present study used molecular docking analyses to assess the molecular interactions and binding affinities of the pyrethroid pesticide etoxazole with essential macromolecules. These include tubulins, DNA topoisomerases, glutamate-1-semialdehyde aminotransferase, glutamate-1-semialdehyde aminotransferase, and protochlorophyllide reductase, which are essential components of the cell machinery that control the dynamics of microtubules, DNA topology, amino acid metabolism, and chlorophyll production, respectively (Table 5). Because of the possible consequences for cellular homeostasis, studying etoxazole interactions with these macromolecules is especially important. Cell division and intracellular trafficking are two examples of the many biological processes that can be affected when tubulins-mediated disruption of microtubule dynamics occurs84. DNA topoisomerase-regulated changes in DNA topology can cause genomic instability and chromosomal abnormalities85.
Additionally, changes in the activity of glutamate-1-semialdehyde aminotransferase can impact the metabolism of amino acids86, and interactions with protochlorophyllide reductase can affect the efficiency of photosynthetic processes and the manufacture of chlorophyll87. Our molecular docking investigation results illustrate the interaction pattern and binding affinities of etoxazole with the selected macromolecules, as shown in Table 5; Fig. 3. These results provide a foundation for understanding the molecular effects of etoxazole and its consequences on chromosomal integrity, photosynthetic efficiency, and cellular functions. Etoxazole showed a strong affinity for the tubulin alpha-1B chain; the free energy of binding was determined to be −7.95 kcal/mol, according to the molecular docking study (Table 5). A value of 1.49 µM was found for the inhibition constant (Ki). Many hydrogen bond formations were present in this interaction, especially involving GLN11 and ALA12. Hydrophobic interactions were crucial in stabilizing the binding, and residues like TYR224 and ALA12 contributed to this process (Fig. 3). With a free energy of −6.43 kcal/mol and an inhibition constant (Ki) of 19.35 µM, etoxazole demonstrated a binding affinity towards the tubulin beta chain (Table 5). Etoxazole formed hydrogen bonds with some amino acids, including HIS227; the number of these interactions with HIS227 was even. Hydrophobic interactions with amino acids such as PHE270, HIS227, ALA231, LEU228, LEU215, PRO358, and ARG359 were observed. Alpha and beta tubulin monomer units combine to form polymeric structures known as microtubules. Microtubules are cytoskeletal components that control cell shape, cell mobility, adhesion, replication, and division as well as directing mitosis and transport inside cells88. Pesticides can directly harm DNA and microtubules. The damage to microtubules has the potential to cause harm to both the cell wall and plasma membrane and may also result in the accumulation of CAs55. The findings of this study indicate that etoxazole may impact microtubule stability, potentially affecting various cellular processes.
DNA topoisomerases are widely distributed enzymes that exhibit both ligase and nuclease functions. These enzymes are vital for several cellular activities, including transcription, replication, DNA duplication, chromatin assembly, and chromosome separation. They also have the ability to control the double helix’s degree of supercoiling, which changes the topological characteristics of DNA89. All varieties of DNA topoisomerases break DNA strands by launching a nucleophilic attack on the phosphodiester backbone of DNA. Type I topoisomerases can cut one of the strands of double-stranded DNA, whereas Type II topoisomerases can cleave both strands of double-stranded DNA molecules, thereby creating double-stranded breaks90. Etoxazole had a strong binding affinity for DNA topoisomerase I, as demonstrated by the molecular docking studies (Table 5). Its estimated free energy was − 7.38 kcal/mol, and its inhibition constant (Ki) was 3.89 µM. Most key binding interactions involved hydrogen bond formation with residues include ARG488 and ALA489. Hydrophobic interactions involving residues ALA489, ARG488, LYS587, ARG590 and ALA586 were also recorded. Etoxazole also showed a strong binding affinity for DNA topoisomerase II, with an inhibitory constant (Ki) of 5.65 µM and a calculated free energy of −7.16 kcal/mol. The creation of hydrogen bonds with amino acids like ASN770 and ARG929, as well as interactions with residues GLU854 and LYS723, were noteworthy binding interactions in this context. This implies that etoxazole may impact DNA topology by altering the activity of DNA topoisomerase I and II. Liman et al.91 suggested that the inhibition of topoisomerase results in the formation of single- and double-strand DNA breaks, impedes DNA replication and diminishes genomic stability. In this case, microtubule damages and disturbance of topoisomerases in etoxazole-treated A. cepa cells may be considered among the causes of genotoxicity together with ROS accumulation and membrane damages.
In photosynthetic plants, glutamate-1-semialdehyde aminotransferase is one of the enzymes involved in the Beale pathway that synthesizes alanine from glutamate92. Etoxazole had a binding affinity of 4.69 µM and a free energy of −7.27 kcal/mol for glutamate-1-semialdehyde aminotransferase (Table 5). Hydrophobic interactions with TYR148, PRO67, VAL245, and LYS271 as well as hydrogen bond interactions with GLY121 and LYS271 were all engaged in the interaction. According to this data, etoxazole may disrupt the activity of glutamate-1-semialdehyde aminotransferase, which might affect metabolic pathways involving amino acids. The inhibition of glutamate-1-semialdehyde aminotransferase has been demonstrated to result in the suppression of alanine synthesis, which in turn affects the biosynthesis of chlorophyll93. Additionally, mutations in glutamate-1-semialdehyde aminotransferase homologue genes in rice were found to cause chlorophyll deficiency92. This suggests that etoxazole-glutamate-1-semialdehyde aminotransferase could contribute to the drop of chlorophyll in the group treated with etoxazole.
In the penultimate stage of the production of chlorophyll, which is necessary for photosynthetic light absorption and energy conversion, protochlorophyllide reductase enzymes catalyze the reduction of protochlorophyllide to chlorophyllide94. This ‘greening’ step is crucial not only for chlorophyll biosynthesis but also for chloroplast development. Therefore, inhibition of this enzyme, defined as a membrane-bound protein with a molecular weight of 37,000 D, has a direct effect on the amount of chlorophyll in the plant95.
With an inhibition constant (Ki) of 385.67 nM and a free energy of binding estimated at −8.75 kcal/mol, etoxazole demonstrated a substantial affinity for protochlorophyllide reductase. Hydrogen bond interactions with VAL14, THY226, GLY13, and VAL223 typified this interaction. The binding was also aided by hydrophobic interactions with TYR219, PHE229, CYS222, VAL223, VAL14, ALA88, and LEU228. According to these results, etoxazole may interfere with the function of protochlorophyllide reductase, which might have an impact on the complex mechanism of chlorophyll production and the linked biological processes involved.
Molecular docking analyses were utilized in this study to investigate the binding interactions between three different DNA molecules (B-DNA dodecamer (1BNA), B-DNA dodecamer D (195D), and DNA (1CP8)) and etoxazole, a pyrethroid pesticide. The results of the docking analysis provide important information on how etoxazole may affect the structure and function of DNA. These results, which are explained in Table 6; Fig. 4, highlight the interactions that etoxazole has, albeit at different affinities, with these DNA molecules. Etoxazole showed affinity with a binding energy of −7.73 kcal/mol and an inhibition constant of 2.14 µM towards the B-DNA dodecamer (1BNA). Nucleotides T8, C9, G10, and C11 in chain A and G16, A17, and A18 in chain B were implicated in these interactions. Etoxazole showed strong affinity for nucleotides A7 and A8 in chain A and T17, T18, and A19 in chain B when it came to B-DNA dodecamer D (195D). This interaction produced a binding energy of −7.88 kcal/mol and an inhibition constant of 1.68 µM. Furthermore, interaction between etoxazole and nucleotide chain 1CP8 occurred with an inhibition constant of 25.41 µM and a binding energy of −6.27kcal/mol. This interaction was mediated through the G4 nucleotides in the A chain of the nucleotide and the C6 and A7 bases in the B chain. Through interactions with both similar and unique strands within DNA structures, the compound’s ability for intercalation has been shown through molecular docking investigations between etoxazole and DNA molecules. Chemicals are stacked between base pairs in DNA molecules during intercalation, a promutagenic phenomenon, but no covalent connections are formed between the chemical agent and the DNA96. Our study is consistent with previous docking studies showing that pesticides can intercalate into DNA molecules97,98. All these results point to the capacity of etoxazole to interact with DNA, perhaps affecting its conformational dynamics and structural stability. The vital role that DNA plays in storing genetic information and coordinating a variety of biological processes may be impacted by these interactions. Additionally, our findings suggest that etoxazole may be able to modify DNA structure by showing a predilection for binding to areas that are rich in particular nucleotide sequences, such as T-C-G-C, G-A-A, A-A, T-T-A, and C-A.
Table 7; Fig. 5 illustrate the damage caused to meristematic cells by etoxazole and the protective effect of A. millefolium. The treatment of AME 1 and AME 2 groups with A. millefolium extract did not result in any observable damage to the meristematic tissue, which was similar to the control group (Fig. 5a-d). The absence of meristematic damage confirms that A. millefolium extract does not exert a toxic effect on A. cepa and this result was consistent with other findings of the study. On the other hand, the administration of etoxazole resulted in the induction of a diverse range of damages to meristematic cells including epidermis cell damage (Fig. 5e), flattened cell nucleus (Fig. 5f), thickening of the cortex cell wall (Fig. 5g) and thickening of the conduction tissue (Fig. 5h). Similarly, Macar et al.8 reported that the application of etoxazole induced a series of meristematic cell damages in A. cepa roots. Moreover, previous studies on the toxicity of pesticides in A. cepademonstrated that pesticides disrupt the structure of root meristematic tissue73,99. The epidermis cells damage and thickening of cortex cell walls may be regarded as a defensive mechanism employed by plants to impede the uptake of etoxazole into root8. While epidermal cells reduce permeability by adopting a more compact and smaller cell structure, the thickening of the cortex cells and conduction tissue with the accumulation of substances such as cellulose and suberin may represent an adaptation to prevent the harmful chemical from reaching the upper parts of the plant via the conduction tissue100. On the other hand, the presence of flattened cell nuclei is indicative of damage to the genetic material, which is corroborated by the genotoxicity results obtained from the present study. When A. millefolium extract and etoxazole were administered together in the ETXAME 1 and ETXAME 2 groups, meristematic cell damages were reduced in a dose-dependent manner (Table 7). A review of the literature revealed no studies demonstrating that A. millefolium mitigates the toxicity associated with pesticides observed in the A. cepa meristem structure. However, previous research has reported that plant extracts with high antioxidant power possess the capacity to prevent meristematic cell damages on root meristem cells in A. cepa72,73,101. The findings of our investigation demonstrated that A. millefolium extract markedly diminished the deleterious effects of etoxazole, particularly oxidative damage, which is the primary cause of impairment to cell membranes and disruption of nuclear structure, ultimately leading to structural disorders of meristematic tissue of roots.
Control (C): Tap water, AME 1: 200 mg/L A. millefolium extract, AME 2: 400 mg/L A. millefolium extract, ETX: 0.5 mL/L etoxazole, ETXAME 1: 0.5 mL/L etoxazole + 200 mg/L A. millefolium extract, ETXAME 2: 0.5 mL/L etoxazole + 400 mg/L A. millefolium extract. ECD: epidermis cell damage, FCN: flattened cell nucleus, TCCW: thickening of the cortex cell wall, TCT: thickening of the conduction tissue. (+++): extremely damaged, (++): moderately damaged, (+): slightly damaged, (-): undamaged.
Etoxazole-induced meristematic cell damage. Normal appearance of epidermis cells (a), normal appearance of cell nucleus-oval (b), normal appearance of cortex cells (c), normal appearance of conduction tissue (d), epidermis cell damage (e), flattened cell nucleus (f), thickened cortex cell wall (g), thickened conduction tissue (h). Bar: 10 μm.
Conclusion
The use of pesticides is crucial factor in increasing agricultural production. However, their widespread use has resulted in a multitude of environmental and public health concerns. The most preferred method to mitigate these adverse effects is use of herbal extracts with high antioxidant power. The present study aimed to investigate the ability of A. millefolium, a plant with well-known therapeutic benefits, to defend against the toxic effects of the hazardous pesticide etoxazole in a multifaceted manner using a eukaryotic plant model. In light of the above, it can be concluded that the A. millefolium extract has effectively mitigated etoxazole-induced phytotoxicity, genotoxicity and oxidative stress in A. cepa. The administration of A. millefolium extract in conjunction with etoxazole was observed to enhance growth, increase MI, increase the levels of both chlorophyll a and chlorophyll b, and reduce the formation of CAs and MNs. Additionally, the extract was found to decrease the levels of MDA, antioxidant enzyme activities, and epidermis cell damage. Molecular docking analysis showed that etoxazole can interact with DNA species, DNA topoisomerases, tubulin proteins, glutamate-1-semialdehyde aminotransferase and protochlorophyllide reductase. This shielding role can be attributed to the high levels of antioxidant phenolic compounds determined within the plant extract. Indeed, A. millefolium extract has such a strong antioxidant effect in A. cepa that its presence reduces both the activity of antioxidant enzymes and membrane damage in etoxazole-induced oxidative stress. Moreover, this study contributed to the elucidation of the mechanism of etoxazole toxicity and offered more proof that A. millefolium had no genotoxic effects. A. millefolium, a plant with a long history of medicinal use, has the potential to serve as a new and reliable pharmacological shield against health problems caused by hazardous pesticides that have emerged in the modern era. It would be useful to encourage the public to use this panacea. Research on the mechanism of action and new ways of use should be continued with different assays. The findings of this study will contribute to the utilization of A. millefolium extract and the advancement of novel pharmaceuticals derived from this extract to mitigate the adverse effects of toxic chemicals.
Statement regarding experimental research on plants
Experimental research and field studies on plants and plant parts (A. cepa bulbs, A. millefolium), including the collection of plant material, comply with relevant institutional, national, and international guidelines and legislation.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Doğanlar, Z. B. et al. Nonoccupational exposure of agricultural area residents to pesticides: pesticide accumulation and evaluation of genotoxicity. Arch. Environ. Contam. Toxicol. 75, 530–544. https://doi.org/10.1007/s00244-018-0545-7 (2018).
Hashimi, M. H., Hashimi, R. & Ryan, Q. Toxic effects of pesticides on humans, plants, animals, pollinators and beneficial organisms. Asian Plant. Res. J. 5, 37–47. https://doi.org/10.9734/aprj/2020/v5i430114 (2020).
El-Sayed, A. & Safar, S. H. Efficacy of Metarhizium anisopliae biopesticide compared with two chitin synthesis inhibitors hexythiazox and etoxazole in Tetranychus Urticae Koch management. Egypt. Acad. J. Biol. Sci. 16, 1–13. https://doi.org/10.21608/EAJBSF.2024.338614 (2024).
Park, W., Lim, W., Park, S., Whang, K. Y. & Song, G. Exposure to etoxazole induces mitochondria-mediated apoptosis in porcine trophectoderm and uterine luminal epithelial cells. Environ. Pollut. 257, 113480. https://doi.org/10.1016/j.envpol.2019.113480 (2020).
Ham, J., You, S., Lim, W. & Song, G. Etoxazole induces testicular malfunction in mice by dysregulating mitochondrial function and calcium homeostasis. Environ. Pollut. 263, 114573. https://doi.org/10.1016/j.envpol.2020.114573 (2020).
Sevgiler, Y., Oruç, E. Ö. & Üner, N. Evaluation of etoxazole toxicity in the liver of Oreochromis niloticus. Pestic Biochem. Phys. 78, 1–8. https://doi.org/10.1016/j.pestbp.2003.09.004 (2004).
Rencüzoğulları, E. et al. The genotoxic effect of the new acaricide etoxazole. Russ J. Genet. 40, 1300–1304. https://doi.org/10.1023/B:RUGE.0000048674.00728.2f (2004).
Macar, O., Kalefetoğlu Macar, T., Çavuşoğlu, K. & Yalçın, E. Risk assessment of oxidative stress and multiple toxicity induced by etoxazole. Sci. Rep. 12, 20453. https://doi.org/10.1038/s41598-022-24966-0 (2022).
Park, H., Lee, J. Y., Park, S., Song, G. & Lim, W. Developmental toxicity and angiogenic defects of etoxazole exposed zebrafish (Danio rerio) larvae. Aquat. Toxicol. 217, 105324. https://doi.org/10.1016/j.aquatox.2019.105324 (2019).
Yilmaz, M., Rencuzogullari, E. & Canli, M. Investigations on the effects of etoxazole in the liver and kidney of Wistar rats. Environ. Sci. Pollut Res. 24, 19635–19639. https://doi.org/10.1007/s11356-017-9601-5 (2017).
Lundstrom, K. Unlocking the therapeutic potential of plant extracts. Future Med. Chem. 8, 245–248. https://doi.org/10.4155/fmc-2015-0012 (2016).
Sun, W. & Shahrajabian, M. H. Therapeutic potential of phenolic compounds in medicinal plants-natural health products for human health. Molecules 28, 1845. https://doi.org/10.3390/molecules28041845 (2023).
Far, B. F., Behzad, G. & Khalili, H. Achillea millefolium: mechanism of action, pharmacokinetic, clinical drug-drug interactions and tolerability. Heliyon 9, e22841. https://doi.org/10.1016/j.heliyon.2023.e22841 (2023).
Okkay, U. et al. Effects of Achillea millefolium on cisplatin induced ocular toxicity: an experimental study. Cutan. Ocul Toxicol. 40, 214–220. https://doi.org/10.1080/15569527.2021.1919137 (2021).
Dias, M. I. et al. Chemical composition of wild and commercial Achillea millefolium L. and bioactivity of the methanolic extract, infusion and decoction. Food Chem. 141, 4152–4160. https://doi.org/10.1016/j.foodchem.2013.07.018 (2013).
Applequist, W. L. & Moerman, D. E. Yarrow (Achillea millefolium L.): a neglected panacea? A review of ethnobotany, bioactivity, and biomedical research. Econ. Bot. 65, 209–225. https://doi.org/10.1007/s12231-011-9154-3 (2011).
Shahani, S. et al. Radioprotective effect of Achillea millefolium L. against genotoxicity induced by ionizing radiation in human normal lymphocytes. Dose Response. 13, 1559325815583761. https://doi.org/10.1177/1559325815583761 (2015).
Adil, M., Dastagir, G., Quddoos, A., Naseer, M. & Filimban, F. Z. HPLC analysis, genotoxic and antioxidant potential of Achillea millefolium L. and Chaerophyllum Villosum Wall ex. Dc. BMC complement. Med. Ther. 24, 91. https://doi.org/10.1186/s12906-024-04344-1 (2024).
Animasaun, D. A. et al. Hazard assessment and cytogenotoxic effect of different concentrations of mercury chloride sterilant using the Allium cepa assay. Discov Toxicol. 1, 2. https://doi.org/10.1007/s44339-024-00002-w (2024).
Silveira, G. L., Lima, M. G. F., Reis, D., Palmieri, G. B., Andrade-Vieria, L. F. & M. J. & Toxic effects of environmental pollutants: comparative investigation using Allium cepa L. and Lactuca sativa L. Chemosphere 178, 359–367. https://doi.org/10.1016/j.chemosphere.2017.03.048 (2017).
Chaleshtori, S. N., Khajoee, M. R., Lorigooini, Z., Anjomshoa, M. & Houshmand, F. Protective effects of Achillea millefolium extract on myocardial infarction induced by isoproterenol in rats. J. Mazandaran Univ. Med. Sci. 32, 1–16 (2022).
Atik, M., Karagüzel, O. & Ersoy, S. Effect of temperature on germination characteristics of Dalbergia sissoo seeds. Mediterr. Agric. Sci. 20, 203–210 (2007).
Staykova, T. A., Ivanova, E. N. & Velcheva, I. G. Cytogenetic effect of heavy-metal and cyanide in contaminated waters from the region of southwest Bulgaria. J. Cell. Mol. Biol. 4, 41–46 (2005).
Fenech, M. et al. HUMN Project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. 534, 65–75. https://doi.org/10.1016/S1383-5718(02)00249-8 (2003).
Drew, H. R. et al. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. 78, 2179–2183. (1981). https://doi.org/10.1073/pnas.78.4.2179
Balendiran, K., Rao, S. T., Sekharudu, C. Y., Zon, G. & Sundaralingam, M. X-ray structures of the B-DNA dodecamer d (CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex. Acta Crystallogr. D Biol. Crystallogr. 51, 190–198. https://doi.org/10.1107/S0907444994010759 (1995).
Katahira, R., Yamashita, Y., Ogawa, H., Yoshida, M. & Katahira, M. Solution structure of the novel antitumor drug UCH9 complexed with d (TTGGCCAA) 2 as determined by NMR. Nucleic Acids Res. 26, 744–755. https://doi.org/10.1093/nar/26.3.744 (1998).
Staker, B. L. et al. The mechanism of topoisomerase I poisoning by a camptothecin analog. PNAS 99, 15387–15392. https://doi.org/10.1073/pnas.242259599 (2002).
Wang, Y. R. et al. Producing irreversible topoisomerase II-mediated DNA breaks by site-specific pt (II)-methionine coordination chemistry. Nucleic Acids Res. 45, 10861–10871. https://doi.org/10.1093/nar/gkx742 (2017).
Lacey, S. E., He, S., Scheres, S. H. & Carter, A. P. Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule. Elife 8, e47145. https://doi.org/10.7554/eLife.47145 (2019).
Mizutani, H. & Kunishima, N. Crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from Aeropyrum pernix. (2011). https://doi.org/10.2210/pdb2ZSL/pdb
Zhang, S. et al. Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574, 722–725. https://doi.org/10.1038/s41586-019-1685-2 (2019).
Guex, N. & Peitsch, M. CSWISS-MODEL and the Swiss‐pdb viewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. https://doi.org/10.1002/elps.1150181505 (2005).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33. https://doi.org/10.1186/1758-2946-3-33 (2011).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. https://doi.org/10.1002/jcc.21256 (2009).
BIOVIA, Dassault Systèmes, B. I. O. V. I. A. D. & Studio San Diego: Dassault Systèmes (2023). (2020).
Kaydan, D., Yagmur, M. & Okut, N. Effects of salicylic acid on the growth and some physiological characters in salt stressed wheat (Triticum aestivum L). J. Agr Sci. 13, 114–119. https://doi.org/10.1501/Tarimbil_0000000444 (2007).
Witham, F. H., Blaydes, D. R. & Devlin, R. M. in Experiments in Plant Physiology. (eds Witham, F. H.) 1–11 (Van Nostrand Reinhold Co, 1971).
Bates, L. S., Waldren, R. P. & Teare, I. D. Rapid determination of free proline for water stress studies. Plant. Soil. 39, 205–207. https://doi.org/10.1007/BF00018060 (1973).
Ünyayar, S., Celik, A., Çekiç, F. Ö. & Gözel, A. Cadmium-induced genotoxicity, cytotoxicity and lipid peroxidation in Allium sativum and Vicia faba. Mutagenesis 21, 77–81. https://doi.org/10.1093/mutage/gel001 (2006).
Zou, J., Yue, J., Jiang, W. & Liu, D. Effects of cadmium stress on root tip cells and some physiological indexes in Allium cepa var. agrogarum L. Acta Biol. Cracov. Ser. Bot. 54, 129–141. (2012). https://doi.org/10.2478/v10182-012-0015-x
Beauchamp, C. & Fridovich, I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. https://doi.org/10.1016/0003-2697(71)90370-8 (1971).
Beers, R. F. & Sizer, I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195, 133–140. https://doi.org/10.1016/S0021-9258(19)50881-X (1952).
Cagri Akman, T. et al. LC-ESI‐MS/MS chemical characterization, antioxidant and antidiabetic properties of propolis extracted with organic solvents from Eastern Anatolia Region. Chem. Biodivers. 20, e202201189. https://doi.org/10.1002/cbdv.202201189 (2023).
Kayir, Ö., Doğan, H., Alver, E. & Bilici, İ. Quantification of phenolic component by LC-HESI-MS/MS and evaluation of antioxidant activities of Crocus ancyrensis (Ankara çiğdemi) extracts obtained with different solvents. Chem. Biodivers. 20, e202201186. https://doi.org/10.1002/cbdv.202201186 (2023).
Lushchak, V. I., Matviishyn, T. M., Husak, V. V., Storey, J. M. & Storey, K. B. Pesticide toxicity: a mechanistic approach. EXCLI J. 17, 1101. https://doi.org/10.17179/excli2018-1710 (2018).
Sun, D., Pang, J., Fang, Q., Zhou, Z. & Jiao, B. Stereoselective toxicity of etoxazole to MCF-7 cells and its dissipation behavior in citrus and soil. Environ. Sci. Pollut Res. 23, 24731–24738. https://doi.org/10.1007/s11356-016-7393-7 (2016).
Çavuşoğlu, K. & Yalçin, E. Spectral shift supported epichlorohydrin toxicity and the protective role of sage. Environ. Sci. Pollut Res. 30, 1374–1385. https://doi.org/10.1007/s11356-022-22288-2 (2023).
Parween, T., Jan, S., Mahmooduzzafar, S., Fatma, T. & Siddiqui, Z. H. Selective effect of pesticides on plant-a review. Crit. Rev. Food Sci. Nutr. 56, 160–179. https://doi.org/10.1080/10408398.2013.787969 (2016).
Tsukagoshi, H. Control of root growth and development by reactive oxygen species. Curr. Opin. Plant. Biol. 29, 57–63. https://doi.org/10.1016/j.pbi.2015.10.012 (2016).
Barut, E. N. et al. Antioxidant capacity, anti-acetylcholinesterase activity and inhibitory effect on lipid peroxidation in mice brain homogenate of Achillea millefolium. Turk. J. Biochem. 42, 493–502. https://doi.org/10.1515/tjb-2017-0084 (2017).
Pereira, J. M. et al. Achillea millefolium L. hydroethanolic extract inhibits growth of human tumor cell lines by interfering with cell cycle and inducing apoptosis. Food Chem. Toxicol. 118, 635–644. https://doi.org/10.1016/j.fct.2018.06.006 (2018).
Ristea, M. E. & Zarnescu, O. Effects of indigo carmine on growth, cell division, and morphology of Allium cepa L. root tip. Toxics 12, 194. https://doi.org/10.3390/toxics12030194 (2024).
Khanna, N. & Sharma, S. Allium cepa root chromosomal aberration assay: a review. Indian J. Pharm. Biol. Res. 1, 105–119. https://doi.org/10.30750/ijpbr.1.3.15 (2013).
Camilo-Cotrim, C. F., Bailão, E. F. L. C., Ondei, L. S., Carneiro, F. M. & Almeida, L. M. What can the Allium cepa test say about pesticide safety? A review. Environ. Sci. Pollut Res. 29, 48088–48104. https://doi.org/10.1007/s11356-022-20695-z (2022).
Pharmawati, M. & Wrasiati, L. P. Chromosomal and nuclear alteration induced by nickel nitrate in the root tips of Allium cepa var. aggregatum. Pollution 9, 702–711. https://doi.org/10.22059/poll.2022.349167.1634 (2023).
Sabeen, M. et al. Allium cepa assay based comparative study of selected vegetables and the chromosomal aberrations due to heavy metal accumulation. Saudi J. Biol. Sci. 27, 1368–1374. https://doi.org/10.1016/j.sjbs.2019.12.011 (2020).
Boumaza, A., Ergüç, A. & Orhan, H. The cytotoxic, genotoxic and mitotoxic effects of Atractylis gummifera extract in vitro. Afr. Health Sci. 24, 295–306. https://doi.org/10.4314/ahs.v24i1.35 (2024).
Seth, C. S., Misra, V., Chauhan, L. K. S. & Singh, R. R. Genotoxicity of cadmium on root meristem cells of Allium cepa: cytogenetic and Comet assay approach. Ecotoxicol. Environ. Saf. 71, 711–716. https://doi.org/10.1016/j.ecoenv.2008.02.003 (2008).
Barman, M. & Ray, S. Cytogenotoxic effects of 3-epicaryoptin in Allium cepa L. root apical meristem cells. Protoplasma 260, 1163–1177. https://doi.org/10.1007/s00709-023-01838-6 (2023).
Kalcheva, V. P., Dragoeva, A. P., Kalchev, K. N. & Enchev, D. D. Cytotoxic and genotoxic effects of Br-containing oxaphosphole on Allium cepa L. root tip cells and mouse bone marrow cells. Genet. Mol. Biol. 32, 389–393. https://doi.org/10.1590/S1415-47572009000200028 (2009).
Prajitha, V. & Thoppil, J. E. Cytotoxic and apoptotic activities of extract of Amaranthus spinosus L. in Allium cepa and human erythrocytes. Cytotechnology 69, 123–133. https://doi.org/10.1007/s10616-016-0044-5 (2017).
Sutan, N. A., Popescu, A., Mihaescu, C., Soare, L. C. & Marinescu, M. V. Evaluation of cytotoxic and genotoxic potential of the fungicide ridomil in Allium cepa L. Analele Stiint ale Univ. Al Cuza Din Iasi. 60, 5–12 (2014).
Zafra-Lemos, L., Cusioli, L. F., Bergamasco, R. & Borin-Carvalho, L. A. De Brito Portela-Castro, A. L. evaluation of the genotoxic and cytotoxic effects of exposure to the herbicide 2, 4-dichlorophenoxyacetic acid in Astyanax lacustris (Pisces, Characidae) and the potential for its removal from contaminated water using a biosorbent. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 865, 503335. https://doi.org/10.1016/j.mrgentox.2021.503335 (2021).
Rasgele, P. G., Demir, F. & Kirankaya, S. G. Determination of micronuclei frequency in Danio rerio for assessing genotoxicity induced by propineb. Drug Chem. Toxicol. 1–6. https://doi.org/10.1080/01480545.2024.2303970 (2024).
Mahapatra, K., De, S., Banerjee, S. & Roy, S. Pesticide mediated oxidative stress induces genotoxicity and disrupts chromatin structure in fenugreek (Trigonella foenum-graecum L.) seedlings. J. Hazard. Mater. 369, 362–374. https://doi.org/10.1016/j.jhazmat.2019.02.056 (2019).
Montaser, S., Mahmoud, M. & Ibrahim, S. Antioxidant capacity of Achillea millefolium L. against cytogenetic and immunological disorders in irradiated human blood cultures. Egypt. J. Radiat. Sci. Appl. 32, 137–144. https://doi.org/10.21608/ejrsa.2019.14648.1077 (2019).
Sule, R. O., Condon, L. & Gomes, A. V. A common feature of pesticides: oxidative stress-the role of oxidative stress in pesticide-induced toxicity. Oxid. Med. Cell. Longev. 5563759. (2022). https://doi.org/10.1155/2022/5563759 (2022).
Dar, M. I., Khan, F. A. & Rehman, F. Responses of antioxidative defense system and composition of photosynthetic pigments in Brassica juncea L. upon imidacloprid treatments. Absjournal 1, 3–15. https://doi.org/10.17582/journal.absjournal/2015/1.1.3.15 (2015).
Üner, N., Oruç, E. & Sevgiler, Y. Oxidative stress-related and ATPase effects of etoxazole in different tissues of Oreochromis niloticus. Environ. Toxicol. Pharmacol. 20, 99–106. https://doi.org/10.1016/j.etap.2004.11.006 (2005).
Kalefetoğlu Macar, T., Macar, O., Yalçιn, E. & Çavuşoğlu, K. Preventive efficiency of cornelian cherry (Cornus mas L.) fruit extract in diniconazole fungicide-treated Allium cepa L. roots. Sci. Rep. 11, 2534. https://doi.org/10.1038/s41598-021-82132-4 (2021).
Yalçin, E. & Çavuşoğlu, K. Spectroscopic contribution to glyphosate toxicity profile and the remedial effects of Momordica charantia. Sci. Rep. 12, 20020. https://doi.org/10.1038/s41598-022-24692-7 (2022).
Yirmibeş, F., Yalçin, E. & Çavuşoğlu, K. Protective role of green tea against paraquat toxicity in Allium cepa L.: physiological, cytogenetic, biochemical, and anatomical assessment. Environ. Sci. Pollut Res. 29, 23794–23805. https://doi.org/10.1007/s11356-021-17313-9 (2022).
Trumbeckaite, S. et al. Achillea millefolium L. s.l. herb extract: antioxidant activity and effect on the rat heart mitochondrial functions. Food Chem. 127, 1540–1548. https://doi.org/10.1016/j.foodchem.2011.02.014 (2011).
Potrich, F. B. et al. Antiulcerogenic activity of hydroalcoholic extract of Achillea millefolium L.: involvement of the antioxidant system. J. Ethnopharmacol. 130, 85–92. https://doi.org/10.1016/j.jep.2010.04.014 (2010).
Sharma, A., Kumar, V., Thukral, A. K. & Bhardwaj, R. Responses of plants to pesticide toxicity: an overview. Planta Daninha. 37, e019184291. https://doi.org/10.1590/S0100-83582019370100065 (2019).
Mishra, K., Sanwal, E., Tandon, P. K. & Gupta, K. Influence of pesticide effluent on Allium cepa L. (onion) plants. Int. J. Environ. 4, 95–105. https://doi.org/10.3126/ije.v4i2.12629 (2015).
Shakir, S. K. et al. Effect of some commonly used pesticides on seed germination, biomass production and photosynthetic pigments in tomato (Lycopersicon esculentum). Ecotoxicol 25, 329–341. https://doi.org/10.1007/s10646-015-1591-9 (2016).
Jan, S., Singh, R., Bhardwaj, R., Ahmad, P. & Kapoor, D. Plant growth regulators: a sustainable approach to combat pesticide toxicity. 3 Biotech. 10, 466. https://doi.org/10.1007/s13205-020-02454-4 (2020).
Yadav, M., Gupta, P. & Seth, C. S. Foliar application of α-lipoic acid attenuates cadmium toxicity on photosynthetic pigments and nitrogen metabolism in Solanum lycopersicum L. Acta Physiol. Plant. 44, 112. https://doi.org/10.1007/s11738-022-03445-z (2022).
Mesquita, A. F., Gonçalves, F. J. & Gonçalves, A. M. The lethal and sub-lethal effects of fluorinated and copper-based pesticides-a review. Int. J. Environ. Res. Public. Health. 20, 3706. https://doi.org/10.3390/ijerph20043706 (2023).
Yang, C. M., Lee, C. N. & Chou, C. H. Effects of three allelopathic phenolics on chlorophyll accumulation of rice (Oryza sativa) seedlings: I. Inhibition of supply-orientation. Bot. Bull. Acad. Sin. 43, 299–304 (2002).
Liu, N. et al. Melatonin alleviates imidacloprid phytotoxicity to cucumber (Cucumis sativus L.) through modulating redox homeostasis in plants and promoting its metabolism by enhancing glutathione dependent detoxification. Ecotoxicol. Environ. Saf. 217, 112248. https://doi.org/10.1016/j.ecoenv.2021.112248 (2021).
de Forges, H., Bouissou, A. & Perez, F. Interplay between microtubule dynamics and intracellular organization. Int. J. Biochem. Cell. Biol. 44, 266–274. https://doi.org/10.1016/j.biocel.2011.11.009 (2012).
Degrassi, F., Fiore, M. & Palitti, F. Chromosomal aberrations and genomic instability induced by topoisomerase-targeted antitumour drugs. Curr. Med. Chem. Anticancer Agents. 4, 317–325. https://doi.org/10.2174/1568011043352920 (2004).
Grimm, B. Primary structure of a key enzyme in plant tetrapyrrole synthesis: glutamate 1-semialdehyde aminotransferase. Proc. Natl. Acad. Sci. 87, 4169–4173. https://doi.org/10.1073/pnas.87.11.416 (1990).
Schulz, R. & Senger, H. Protochlorophyllide reductase: a key enzyme in the greening process in Pigment-protein Complexes in Plastids: Synthesis and Assembly (eds Sundqvist, C. & Ryberg, M.) 179–218 (Academic Pres Inc., (1993).
Xu, Q. Q. et al. The expression and potential role of tubulin alpha 1b in Wilms’ tumor. Biomed Res. Int. 9809347. (2020). https://doi.org/10.1155/2020/9809347 (2020).
Mastrangelo, S. et al. The DNA-topoisomerase inhibitors in cancer therapy. Biomed. Pharmacol. J. 15, 553–562. https://doi.org/10.13005/bpj/2396 (2022).
Talukdar, A. et al. I inhibitors: challenges, progress and the road ahead. Eur. J. Med. Chem. 236, 114304. https://doi.org/10.1016/j.ejmech.2022.114304 (2022).
Liman, R., Ali, M. M., Istifli, E. S., Ciğerci, İ. H. & Bonciu, E. Genotoxic and cytotoxic effects of pethoxamid herbicide on Allium cepa cells and its molecular docking studies to unravel genotoxicity mechanism. Environ. Sci. Pollut Res. 29, 63127–63140. https://doi.org/10.1007/s11356-022-20166-5 (2022).
Jiang, M. et al. An alanine to valine mutation of glutamyl-tRNA reductase enhances 5-aminolevulinic acid synthesis in rice. Theor. Appl. Genet. 135, 2817–2831. https://doi.org/10.1007/s00122-022-04151-7 (2022).
Zhao, Y. et al. Effect of low temperature on chlorophyll biosynthesis and chloroplast biogenesis of rice seedlings during greening. Int. J. Mol. Sci. 21, 1390. https://doi.org/10.3390/ijms21041390 (2020).
Vedalankar, P. & Tripathy, B. C. Evolution of light-independent protochlorophyllide oxidoreductase. Protoplasma 256, 293–312. https://doi.org/10.1007/s00709-018-1317-y (2019).
Begley, T. P. & Young, H. Protochlorophyllide reductase. 1. Determination of the regiochemistry and the stereochemistry of the reduction of protochlorophyllide to chlorophyllide. J. Am. Chem. Soc. 111, 3095–3096. https://doi.org/10.1021/ja00190a071 (1989).
Seven, B., Kültiğin, Çavuşoğlu, Yalçin, E. & Acar, A. Investigation of cypermethrin toxicity in Swiss albino mice with physiological, genetic and biochemical approaches. Sci. Rep. 12, 11439. https://doi.org/10.1038/s41598-022-15800-8 (2022).
Kuloğlu, S. S., Yalçin, E., Çavuşoğlu, K. & Acar, A. Dose-dependent toxicity profile and genotoxicity mechanism of lithium carbonate. Sci. Rep. 12, 13504. https://doi.org/10.1038/s41598-022-17838-0 (2022).
Onur, B., Çavuşoğlu, K., Yalçin, E. & Acar, A. Paraquat toxicity in different cell types of Swiss albino mice. Sci. Rep. 12, 4818. https://doi.org/10.1038/s41598-022-08961-z (2022).
Çakir, F., Kutluer, F., Yalçin, E., Çavuşoğlu, K. & Acar, A. Deep neural network and molecular docking supported toxicity profile of prometryn. Chemosphere 340, 139962. https://doi.org/10.1016/j.chemosphere.2023.139962 (2023).
Yalçın, E., Uzun, A. & Çavuşoğlu, K. In vivo epiclorohidrine toxicity: cytogenetic, biochemical, physiological, and anatomical evidences. Environ. Sci. Pollut Res. 26, 22400–22406. https://doi.org/10.1007/s11356-019-05518-y (2019).
Himtaş, D., Yalçin, E., Çavuşoğlu, K. & Acar, A. In-vivo and in-silico studies to identify toxicity mechanisms of permethrin with the toxicity-reducing role of ginger. Environ. Sci. Pollut Res. 31, 9272–9287. https://doi.org/10.1007/s11356-023-31729-5 (2024).
Alara, O. R., Abdurahman, N. H. & Ukaegbu, C. I. Extraction of phenolic compounds: a review. Curr. Res. Food Sci. 4, 200–214. https://doi.org/10.1016/j.crfs.2021.03.011 (2021).
Stojković, D. et al. In situ antioxidant and antimicrobial activities of naturally occurring caffeic acid, p-coumaric acid and rutin, using food systems. J. Sci. Food Agric. 93, 3205–3208. https://doi.org/10.1002/jsfa.6156 (2013).
Khan, M. S., Abul Qais, F., Ahmad, I., Hussain, A. & Alajmi, M. F. Genotoxicity inhibition by Syzygium cumini (L.) seed fraction and rutin: understanding the underlying mechanism of DNA protection. Toxicol. Res. 7, 156–171. https://doi.org/10.1039/c7tx00269f (2018).
Abou Baker, D. H. Achillea millefolium L. ethyl acetate fraction induces apoptosis and cell cycle arrest in human cervical cancer (HeLa) cells. Ann. Agric. Sci. 65, 42–48. https://doi.org/10.1016/j.aoas.2020.03.003 (2020).
Salomon, L. et al. Comparison of the phenolic compound profile and antioxidant potential of Achillea Atrata L. and Achillea millefolium L. Molecules 26, 1530. https://doi.org/10.3390/molecules26061530 (2021).
Radušienė, J. et al. Trends in phenolic profiles of Achillea millefolium from different geographical gradients. Plants 12, 746. https://doi.org/10.3390/plants12040746 (2023).
Prasad, R. & Prasad, S. B. Modulatory effect of rutin on the antitumor activity and genotoxicity of cisplatin in tumor-bearing mice. Adv. Pharm. Bull. 11, 746–754. https://doi.org/10.34172/apb.2021.084 (2021).
Salau, V. F. et al. Vanillin and vanillic acid modulate antioxidant defense system via amelioration of metabolic complications linked to Fe2+-induced brain tissues damage. Metab. Brain Dis. 35, 727–738. https://doi.org/10.1007/s11011-020-00545-y (2020).
Tai, A., Sawano, T., Yazama, F. & Ito, H. Evaluation of antioxidant activity of vanillin by using multiple antioxidant assays. Biochim. Biophys. Acta - Gen. Subj. 1810, 170–177. https://doi.org/10.1016/j.bbagen.2010.11.004 (2011).
Al-Baqami, N. M. & Hamza, R. Z. Synergistic antioxidant capacities of vanillin and chitosan nanoparticles against reactive oxygen species, hepatotoxicity, and genotoxicity induced by aging in male Wistar rats. Hum. Exp. Toxicol. 40, 183–202. https://doi.org/10.1177/0960327120943267 (2021).
Sefi, M. et al. Beneficial role of vanillin, a polyphenolic flavoring agent, on maneb-induced oxidative stress, DNA damage, and liver histological changes in Swiss albino mice. Hum. Exp. Toxicol. 38, 619–631. https://doi.org/10.1177/0960327119831067 (2019).
Song, X., Wang, Y. & Gao, L. Mechanism of antioxidant properties of quercetin and quercetin-DNA complex. J. Mol. Model. 26, 133. https://doi.org/10.1007/s00894-020-04356-x (2020).
Darband, S. G. et al. Quercetin attenuated oxidative DNA damage through NRF2 signaling pathway in rats with DMH induced colon carcinogenesis. Life Sci. 253, 117584. https://doi.org/10.1016/j.lfs.2020.117584 (2020).
Manuja, R., Sachdeva, S., Jain, A. & Chaudhary, J. A comprehensive review on biological activities of p-hydroxy benzoic acid and its derivatives. Int. J. Pharm. Sci. Rev. Res. 22, 109–115 (2013).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Z.Ş.T, T.K.M., O.M., E.Y. and K.Ç. designed the experiments; Z.Ş.T, T.K.M., O.M., E.Y., K.Ç, A.A. and F.K. performed the analyses; K.Ç. carried out the statistical analysis; Z.Ş.T., T.K.M. and O.M. wrote the manuscript with the help of E.Y., K.Ç., A.A. and F.K.; T.K.M. edited the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Correspondence and requests for materials should be addressed to T.K.M.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Topatan, Z.Ş., Kalefetoğlu Macar, T., Macar, O. et al. Alleviatory efficacy of achillea millefolium L. in etoxazole-mediated toxicity in allium cepa L. Sci Rep 14, 31674 (2024). https://doi.org/10.1038/s41598-024-81586-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-81586-6
Keywords
This article is cited by
-
Alchemilla vulgaris cytoprotection against methyl methanesulfonate by experimental and molecular docking approaches
Scientific Reports (2026)
-
Quantitative phenolic profiling and protective effects of grape seed extract on mancozeb-induced cellular and genetic toxicity
Scientific Reports (2025)
-
The natural and eco-friendly role of Cassia angustifolia in reducing carbaryl toxicity at environmentally relevant concentration
Scientific Reports (2025)
-
Integrated evaluation of methidathion-induced toxicity in Allium cepa with a multifactorial biological approach
Scientific Reports (2025)
-
Synergistic and antagonistic contributions of main components to the bioactivity profile of Anethum graveolens extract
Scientific Reports (2025)