Abstract
One of the most fatal diseases that affect people is skin cancer. Because nevus and melanoma lesions are so similar and there is a high likelihood of false negative diagnoses challenges in hospitals. The aim of this paper is to propose and develop a technique to classify type of skin cancer with high accuracy using minimal resources and lightweight federated transfer learning models. Here minimal resource based pre-trained deep learning models including EfficientNetV2S, EfficientNetB3, ResNet50, and NasNetMobile have been used to apply transfer learning on data of shape\(\:\:224\times\:224\times\:3\). To compare with applied minimal resource transfer learning, same methodology has been applied using best identified model i.e. EfficientNetV2S for images of shape\(\:\:32\times\:32\times\:3\). The identified minimal and lightweight resource based EfficientNetV2S with images of shape \(\:32\times\:32\times\:3\) have been applied for federated learning ecosystem. Both, identically and non-identically distributed datasets of shape \(\:32\times\:32\times\:3\) have been applied and analyzed through federated learning implementations. The results have been analyzed to show the impact of low-pixel images with non-identical distributions over clients using parameters such as accuracy, precision, recall and categorical losses. The classification of skin cancer shows an accuracy of IID 89.83% and Non-IID 90.64%.
Similar content being viewed by others
Introduction
According to the “World Health Organization (WHO),” one-third of cancer cases worldwide are caused by skin cancer, the deadliest kind of malignant cell. It is brought on by the fast growth of aberrant skin cells that result from genetic deformities or alterations in faulty “Deoxyribonucleic acid (DNA)”1. Ecological and hereditary variables like extended UV radiation exposure—specifically, UV-A, which has a long wavelength, and UV-B, which has a short wavelength—can cause skin cancer by causing the skin’s pigment-producing cells, melanocytes, to develop uncontrollably. Skin carcinogenic cells that are frequently documented include “Squamous Cell Carcinoma (SCC)”, “Basal Cell Carcinoma (BCC)”, and “Malignant Melanoma (MM)”2. Out of all these different skin lesions, malignant melanoma is the deadliest with the highest number of incidences. SCC and BCC, on the other hand, are non-melanocytic malignancies that are considered a benign. The National Skin-Cancer Institute estimates that skin cancer is the most common diagnosis, with most occurrences occurring in the United States. Skin illnesses come in different forms, including SCC melanoma, intraepithelial carcinoma, and BCC. According to reports, melanoma has the highest death rate among skin malignancies, with a rate of 1.68%3. However, although BCC is not frequently deadly, it is the most prevalent type of skin cancer and has a significant negative impact on healthcare resources4.
Many of the diseases are curable if they are discovered early on, before they have had a chance to spread. Specialists use a technique called dermoscopy, which uses polarization to lessen surface reflection and strong light to observe changes in the skin5. Accurately diagnosing a skin condition is difficult, though, because numerous visual cues must be used to aid in the diagnosis, including the shape of each lesion, the distribution of body sites, color, scaling, and arrangement of lesions. The diagnostic procedure can get complicated if the constituent parts are examined individually6. For example, the ABCD principles, pattern analysis, Menzies approach, and 7-Point Checklist are the four main clinical diagnosis techniques for melanoma. Using these techniques, skilled doctors are typically the only ones who can obtain good diagnosis accuracy7.
Dermatologists have noted that the visual similarities between a melanoma lesion and a mole, pigmentation, or non-melanocytic lesions might make diagnosis challenging. Malignant lesions are lumps with an uneven and amoeboid surface that grow quickly8. They are usually larger than 5 mm, asymmetrical in shape, and can have deep gray, black, or brown hues. A lesion can be visually identified as a cluster of different colored contours. The melanoma lesion may develop quickly and cause bleeding, ulceration, irritation, and inflammation. The dermatologists concur, however, a lesion from melanoma may not exhibit any systemic symptoms and may spread to any part of the body regardless of exposure to direct sunlight9. Traditionally, skin cancer has been diagnosed and identified through physical screening and ocular examination of lesions since the early 1900s. Dermatologists use observations of modifications in shape, colour, or dimension. Because of the visual complexity of skin lesions, these standard procedures are complicated, prone to error, and time-consuming. For a precise identification of the lesion during physical examination, a qualified professional is required. As a result, non-invasive techniques have grown in importance over time, and more frequently than not, current, affordable tools like dermoscopy and epiluminescence microscopy are used, producing results with higher accuracy than those of earlier techniques10. Dermoscopic, on the other hand, is a far more effective tool for lesion detection. To improve the identified stain lucidity and reduce appearance on the surface of skin, dermoscopic equipment is utilized to enlarge as well as highlight the visuals of the affected area of skin. Using these contemporary dermoscopy instruments greatly enhances diagnosis performances.
Sensitive patient data never leaves the original place thanks to federated learning, which enables deep learning models to be trained directly on decentralized data sources (such hospital or clinical databases). Since just the model parameters are disclosed, there is less chance of personal information being re-identified or exposed—especially in unusual circumstances like uncommon skin malignancies. Federated learning reduces the possibility of data breaches that may arise from centralized data collecting and storage by storing the data locally. Hackers have a harder time obtaining critical patient data since there is a smaller attack surface because data never leaves the local area.
Hence, the major contributions in this paper are as follows-.
-
1.
To propose and develop a technique to classify type of skin cancer with high accuracy using minimal resources and lightweight federated transfer learning models.
-
2.
Reduce resource utilization by implementing pre-trained deep learning models with reduced shape datasets using EfficientNetV2S, EfficientNetB3, ResNet50, and NasNetMobile and compare with the transfer learning approaches using same algorithms.
-
3.
The identified minimal and lightweight resource based EfficientNetV2S have been applied for federated learning ecosystem in both IID and Non-IID datasets.
Rest of the paper is organized as follows: In “Related work” section the literature study related to skin cancer detection is explored using different techniques. The materials used for proposing the model is explain in “Materials and methods” section evaluate result based on different terms is discussed in “Result and discussion” section and shows comparison with other state-of-the-art models in “Discussion” section. The last “Conclusion” section shows the conclusion part with some future scope.
Related work
One kind of skin cancer that can cause malignant tumors on the skin is melanoma. Dermatological photos are used to detect skin cancer. A survey of various sophisticated machine learning algorithms for diagnosing skin cancers was proposed by Bhatt et al.11. After gathering data from several research, the performance of support vector machines, k-nearest neighbors, and convolutional neural networks on comparative datasets was examined. A deep learning-based deep convolutional neural network (DCNN) model for the accurate classification of benign and malignant skin lesions was presented by Ali et al.12. To evaluate its performance, our proposed DCNN model is compared with several transfer learning models, such as AlexNet, ResNet, VGG-16, DenseNet, MobileNet, etc. Salem et al.13 provide a two-phase method for dividing tumors in photos into benign and malignant categories. The first step involves using an image processing-based technique to extract a mole’s diameter, color variation, border irregularity, and asymmetry. Using a genetic algorithm, lesions are classified in the second step.
Ilkin et al.14 created a hybrid classification algorithm by combining the SVM algorithm with a heuristic optimization technique. This technique uses the Bacterial Colony algorithm (hybSVM) to improve the SVM algorithm, which uses a Gaussian Radial Basis Function (RBF). The model was validated using 10 cross-fold validation on two distinct datasets, PH2 and ISIC. Based on data from PH2 and ISIC, the AUC value was 97%, 98%, and the operation time was 11.9,26.5 s, respectively. Li et al.15 examine the research from the perspectives of the type of disease, data collection, data processing, data augmentation, deep learning framework, model performance, and picture recognition model for skin diseases. Additionally, we provide an overview of the conventional and machine learning approaches for diagnosing and treating skin diseases. In addition, we assess the state of this field’s development and forecast four future research possibilities. Hosny et al.16 provide a method for automatically classifying skin lesions that has a higher classification rate by utilizing a pre-trained deep neural network and the transfer learning theory. The Alex-net has seen several applications of transfer learning, including weight optimization and the replacement of a softmax layer—which can handle two or three different types of skin lesions—for the classification layer, and expanding the dataset by adding both random and fixed rotation angles. The new softmax layer may classify the segmented colour picture lesions as nevus, melanoma, or melanoma, seborrhoeic keratosis, and nevus.
Patil and Bellary17. discussed about the tumor thickness or cancer stage are the primary factors that determine a patient’s diagnosis at the time of surgery. Bardou et al.18 examine two machine learning techniques for the automatic classification of histological photos related to breast cancer into two categories: malignant and benign, as well as sub-classes of each. In the first technique, a set of handcrafted features is extracted and trained using support vector machines using two coding models (locality limited linear coding and bag of words). In the second approach, convolutional neural networks are designed. The afflicted lesions were isolated by Ahammed et al.19 using the automatic Grabcut segmentation approach. We utilize statistical features and Gray Level Co-occurrence Matrix (GLCM) approaches to effectively categorize the skin photographs based on the collected data to identify underlying input patterns from the skin pictures. Table 1 displays some of the findings from various authors’ use of pre-trained and hybrid deep learning models to detect skin cancer.
With types of skin cancer dataset some authors conduct detection and classification experiment using different deep learning models at an early stage. Authors used ISIC dataset and some use HAM10000 dataset with DermNet dataset. With pre-trained models the authors detect lesions with an accuracy of 93%. Some authors combine the multimodal transformers with deep learning models and detect a lesion classification accuracy of 92%. Naeem et al.28 propose SNC_Net, which combines “Handmade (HC)” feature extraction techniques with DL models to combine features extracted from dermoscopic images to enhance the classifier’s performance. For classification, a CNN is utilized. With a precision of 98.31%, recall of 97.89%, accuracy of 97.81%, and F1 score of 98.10%, the suggested model performed better than the four baseline models and the SOTA classifiers. Naeem and Anees29. introduced DVFNet, a deep learning-based technique for identifying skin cancer from dermoscopy pictures. Images are pre-processed using anisotropic diffusion techniques to reduce noise and artifacts, improving the quality of the images for the purpose of detecting skin cancer. This study uses the Histogram of Oriented Gradients (HOG) in conjunction with the VGG19 architecture to extract discriminative features.
Riaz et al.30 study investigates the true positive rate (TPR), true negative rate (TNR), area under the curve (AUC), and accuracy (ACC) of the FL and TL classifiers by comparing them to the performance measures documented in research papers. Naeem et al.31 The suggested model, called SCDNet, classifies different forms of skin cancer by combining convolutional neural networks (CNN) with Vgg16. Additionally, the suggested method’s accuracy is contrasted with that of the four cutting-edge pre-trained classifiers in the medical field, Resnet 50, Inception v3, AlexNet, and VGG19. Naeem and Anees32. paper demonstrates a recently created deep learning model that utilizes ResNet101 and Xception, two cutting edge AI methods. When borderline SMOTE is used, performance is significantly improved. The four benchmark classifiers and the suggested technique are compared. The prediction accuracy of the X_R101 model is 98.21%. Dermatologists and other medical professionals’ benefit from the method’s efficacy and accuracy in early detection of skin cancer.
Naeem et al.33 compare the gene expressions of local and metastatic prostate cancer, as well as the differentially expressed genes (DEGs) and biochemical pathways linked to the development of prostate cancer metastases, machine learning is used to uncover possible biomarkers. Ayesha et al.34 proposed a new deep learning architecture is put forth to solve these problems, which include low accuracy, deployment on edge devices, computational expenses, long execution durations, and the persistence of manual feature extraction processes. Using a SoftMax activation function in the last dense layer, a composite feature vector created by these CNN models is then used to classify skin cancer. Our proposed federated learning approach used five different deep learning models with nine different skin cancer datasets for the detection of lesions at an early stage.
Materials and methods
For the identification of skin Melanoma lesion classification, the research approach consisted of three basic procedures. The data were first labeled, resized to two different sizes, and saved in JPG format with a \(\:224\times\:224\) and \(\:32\times\:32\) size to pre-process the data35. Next, divide the dataset in half. Subsequently, eight different transfer learning models and minimal memory pre-trained models were trained, and the results were analyzed. Performance measures were reviewed to identify the best performing framework. Third, “Federated Learning (FL)” was implemented using both “Independent Identically Distributed (IID)” and “Non-Independent Identically Distributed (Non-IID)” datasets. For this, it was necessary to create both client websites and a central server. The median model’s performance was compared with the outcomes of the FL approach, and the overall model was selected by the federated technique using a voting aggregation mechanism.
Data gathering and pre-processing
The skin lesions are gathered from publicly available resource (https://www.kaggle.com/datasets/andrewmvd/isic-2019). The total number of Melanoma skin lesion photos in the study’s dataset is 25,331. Figure 1 shows nine different forms of skin illnesses along with a class name.
The collection only includes JPG-formatted photos with proper labels. Figure 2 shows a breakdown of the image distribution. The 80:20 was used to divide the dataset into training and testing datasets to mimic a federated environment. During the dataset’s preparation, the pictures were converted to grayscale and shrunk to a standard pixel size of \(\:224\times\:224\) and\(\:32\times\:32\). The training subset is given to the model to aid it in learning the intricate details of the images.
In contrast, the subsets used for testing and training are separated. By feeding it to the model at the conclusion of each epoch and assessing its output, it is used to track the model’s performance. After training is finished, the test subset on new data is used to evaluate the model’s overall performance. Table 2 displays the appropriate values for each of the employed augmentation strategies36. This pre-processing step was necessary to make sure that it will work with the requirements of the DL model. The parts belonging to N clients were then randomly selected from the pre-processed data. Every customer was provided with pre-processed data that was utilized by them for training and FL process assessment. The distribution of the data was designed to simulate a scenario where every user was given access to a subset of the dataset.
Federated learning
“Federated Learning (FL)” is a new technique for machine learning which aims to solve the problems caused by restrictive confidentiality of data regulations as well as a dearth of dataset that is easily accessible. A centralised server and a customer are involved37,38,39. Training models without sending the real data is made possible by FL, which uses decentralized end devices and local servers. The central server receives updated weights from the local models that have been trained on customer datasets, protecting the anonymity required for medical data diagnosis. A FL system consists of three primary components: the clients, the server, and the communication configuration. According to the FL’s specified framework, the system consists of M clients (\(\:{M}_{1},\)\(\:{M}_{2},\:{M}_{3},\dots\:\dots\:{M}_{n}\)), each having their own datasets (\(\:{D}_{1},\:{D}_{2},\dots\:\dots\:{D}_{n}\)),). The complete dataset, designated as\(\:{D}_{n}\), is formed by joining individual client datasets (\(\:{D}_{1},\:{D}_{2},\dots\:\dots\:\dots\:.{D}_{n}\)).
In the context of FL, the phrases “IID” and “non-IID” relate to the data distributions among participating clients. When employing IID data in FL, the dispersal of data among all users or devices is similar31. The data from each client are unique and have the same statistical distribution. In the context of picture classification, for example, as per the IID dataset, every user has a comparable proportion of images from various classifications. In a non-IID scenario, the dispersal of data amongst user is neither independent nor similar, in real time scenarios, the availability of data at different locations will be different. To simulate such environment, analysis over Non-IID dataset have been implemented in this paper.
Proposed federated transfer learning model
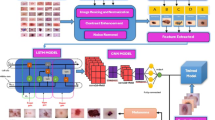
This paper is majorly focused on to develop a lightweight transfer learning model to detect skin cancer in privacy preserved federated learning ecosystem for 9 mentioned classes. Figure 3, initially representing the role of actual image size i.e. \(\:224\times\:224\times\:3\) in comparison to the reduced image size i.e. \(\:32\times\:32\times\:3\) for the training of lightweight transfer learning models including “EfficientNetV2S”, “MobileNetV2”, “EfficientNetB3”, “ResNet50”, and “NasNetMobile”. EfficientNetV2 as a best trained model get identified for the data images of size\(\:224\times\:224\times\:3\). Further same EfficeintNetV2S has been applied to reduced size image data of \(\:32\times\:32\times\:3\) and compared its results with earlier image size data results.
Further, identified model EfficientNetV2S with data size of \(\:32\times\:32\times\:3\) has been recommend using in federated learning ecosystem to achieve the implementation of lightweight federated transfer learning. Here in Fig. 4, four federated learning clients have been deputed along with one federated server to achieve required simulation. Each client is pre-processing its images to reduce size and apply training and validation of local model using pre-trained EfficientNetV2S.
The local trained model sent to the federated server to make federated averaged model and same averaged global model sent back to each client for the updates of the weights of available local model. This process repeated as per number of communication rounds for global updates and number of epochs for local updates.
Result and discussion
The results obtained using the suggested procedures are presented and analyzed in this section. Using a dataset on skin cancer, this section presents a thorough explanation of the evaluation’s “Accuracy”, “Precision”32, “Recall Rate”, and “Loss”40,41,42. The current section discusses a comparative examination of suggested datasets using various deep learning algorithms.
Model training
For this experiment, a Windows 10 PC, a Jupyter notebook, a 64-bit operating system, and 8 GB of Google Drive storage were utilized. Both the Tensorflow backend and the Keras 2.4.3 framework were utilized to facilitate the training and validation of the deep neural network. The evaluation stage, which makes it possible to calculate the difference between the predicted and actual value, is an essential part of the suggested model. These were created to categorize a significant number of classes that had nothing to do with our research. After that, each pooled feature map was compressed, and max pooling layers were added to reduce the size of the feature maps. The dropout layer was inserted after the flattened layer to avoid overfitting.
Metrics
All of the methods’ outcomes were integrated into an exceptional matrix notation that included multiple measurements, such as “accuracy,” “precision,” “recall,” and “F1-score.” The accuracy43 measures how well a neural network can identify a goal with total positive values given the degree of positive values displayed in Eq. 1. Equation 2 shows that the recall value44,45 is dependent on how successfully a neural network associates a goal with the total value.
Two definitions of correctness are as follows: either the accurate neural community with positive test detection identified by Eq. 3, or the difference between the actual and anticipated results46.
Equation 4 represents the F1-score47, which is utilized to calculate the mean precision and recall value with the total value.
Baseline result of different models
This section covers the execution assessment of transfer learning techniques and traditional CNN techniques to diagnose the cutaneous melanoma illness. Comparisons with limited memory pre-trained models include transfer learning techniques “EfficientNetB3”, ”VGG16”, ”ResNet152V2”, ”VGG19”, ”InceptionResNetV2”, ”MobileNetV2”, ”DenseNet201”, ”Xception”. Different characteristics included the precision, recall rate, loss, and training-validation accuracy are calculated. The Table 3 shows result of skin lesion with size of \(\:224\times\:224.\) Before augmentation the EfficientNetB3 shows an accuracy of 87.86% and validation accuracy of 69.07% which is better in comparison with DenseNet201, EfficientNetV2S, MobileNetV2, VGG16, and VITB16. The EfficientNetB3 achieved validation accuracy of 72.39% with loss of 1.48.
Figure 5 shows different evaluation result using DenseNet201, EfficientNetB3, EfficientNetV2S, MobileNetV2, VGG16, and VITB16 models. It is clearly visible from the graphs that the EfficientNetB3 shows maximum accuracy more than 85% with a loss of 0.69.
Figure 5 shows the validation result of different models on different parameters such as validation accuracy, validation precision, validation loss, and validation recall. From the graphs the EfficientNetB3 shows a validation accuracy of 69.07% which is far better than other models. The validation precision of EfficientNetB3 is 72.39% with validation recall of 64.41%. The result with VGG16 shows up and down after every 5th epoch. The result of all the models is shown using 20 epochs. The validation accuracy with VGG16 drops down and reaches maximum value of 42%. The result after data augmentation is shown in Table 4. The model EfficientNetB3 shows an accuracy of 93.22% with validation accuracy of 94.74%. The data after augmentation of size \(\:224\times\:224\) shows minimum loss of 0.51 with VITB16 model. But the EfficientNetB3 shows a loss of 0.58. As clearly shown in Fig. 6 the graphical representation of all the models.
In Fig. 6 the EfficientNetB3 shows model accuracy of 93.22% after epoch 10 and still constant till epoch 20. With VITB16 the accuracy increases once reaches 60% and then increases slowly till epoch 20. The VGG16 shows least validation accuracy after augmentation of 13.33% with zero precision and recall value and with highest loss value of 2.21.
The validation loss with VGG16 reaches maximum with the loss of data or objects part. In comparison EfficientNetB3 and VITB16 shows minimal validation loss with values of 0.58 and 0.51 respectively.
Baseline data results with minimal memory models
The baseline result with minimal memory based pre-trained models including ‘EfficientNetV2S’, ‘MobileNetV2’,‘EfficientNetB3’,‘ResNet50’,‘NASNetMobile’45. With minimal memory and image size of \(\:224\times\:224\) the Fig. 7 shows the result in which maximum validation accuracy of 55.08% is achieved with EfficientNetV2S with validation precision of 56.68%. As the memory size is less and the image size is more the models do not detect the lesions object with good accuracy.
With 20 epochs the loss 0.65 reaches maximum with MobileNetV2, the minimum loss of 1.87 is shown with ResNet50 with an accuracy of 57.20%. Each epoch models performance is evaluated and recorded in graph 6. The result with minimal memory and size \(\:32\times\:32\) is shown in Fig. 8 with augmented dataset.
The minimal memory model result with 20 epochs is shown in Table 5 based on different evaluation metrics. With dataset of size \(\:32\times\:32\)the detection of objects with EfficientNetV2S reaches maximum accuracy of 91.67% with a validation precision of 92.36% and loss of 0.41.
Based on different evaluation metrics, the EfficientNetV2S shows maximum accuracy of 91.67% with validation precision of 92.36%. The result in terms of confusion metric is shown in Fig. 9. The confusion metric of \(\:32\times\:32\times\:3\) image size is shown in Fig. 9. The left side column shows metric for non-augmented dataset and right side shows metric for augmented data.
Table 6 shows the data outcome using federated learning IID data and Non-IID data with image size of \(\:32\times\:32\times\:3\) with augmented images only. Using IID data the accuracy achieved is 98.17% which is better in comparison with all other data type.
The training and validation result of clients using federated learning models is shown in Fig. 10.
The training result with Non-IID data shows an accuracy of 97.63% with precision of 97.90% and validation accuracy using Non-IID is 90.64% with loss of 0.41 which is more in comparison with all other models as shown in Fig. 11. The training accuracies in both IID and non-IID datasets hold steady across several clients, suggesting that performance indicators do not significantly change during the training phase. On the other hand, there are slight variations in performance indicators between various customers when it comes to validation findings.
Discussion
The performance of the proposed model is better than that of the other pertinent research. In this section, we compared our proposed model with approaches that have been used in the past for the classification of photographs of skin lesions. Additionally, we presented the outcomes of using FL on both IID and non-IID datasets. Various pre-trained models were utilized to classify images of skin conditions while protecting privacy of the data. Table 7 shows the comparison of our proposed model with other models. The researchers used U-Net model for the segmentation of lesions at an early stage and then classify with CNN classifier.
In our work, the federated learning also introduced with the deep learning models. In which the client-side data is collected with IID and Non-IID dataset. The work shows validation accuracy of 89.83% and 90.64% with IID and Non-IID. The strength of this paper is proposing emerging technique of feature extraction and selection using pretrained deep learning models which dramatically reduce the resource requirement in comparison to the transfer learning approaches without reducing classification accuracy and other metrics. Paper also explains the impact of non-IID distribution of utilized dataset on proposed methodology, where least impact has been noticed in comparison to IID dataset. The limitation of the proposed methodology is to deploy in a real time ecosystem using mobile or handheld device. Same we have considered for our future research.
Conclusion
Melanoma is a deadly type of skin cancer that causes six out of every seven deaths from skin cancer. The main aim of this paper was to provide a data privacy preserved technique to classify type of skin cancer with high accuracy using minimal resources over federated transfer learning models. The applied minimal resource based best identified pretrained deep learning model i.e. EfficientNetV2S with images of shape \(\:32\times\:32\times\:3\) have resulted better results in comparison to the EfficientNetV2S, EfficientNEtB3, ResNEt50, and NasNetMobile with image data of shape \(\:224\times\:224\times\:3\). The identified minimal and lightweight resource based EfficientNetV2S with images of shape \(\:32\times\:32\times\:3\) have been applied for federated learning ecosystem and identified with the same outcomes in comparison to single system implementations. Both, identically and non-identically distributed datasets of shape \(\:32\times\:32\times\:3\) have been analyzed as a better approach with both identical and non-identical distributions. In conclusion, the applied federated learning has resulted required parametric convergence in both identical and non-identical data distributions. Similar could be used for future implementations for real time applications. As future work, the model’s integration into clinical workflows for early skin cancer screening will be explored, focusing on its potential to improve patient outcomes. Further, the pilot studies or clinical trials will be necessary to validate its effectiveness in real-world applications.
Data availability
The data that support the findings of this study are available on request from the first author i.e. Vikas Khullar.
References
Goyal, M., Knackstedt, T., Yan, S. & Hassanpour, S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities. Comput. Biol. Med. 127, 104065 (2020).
Chang, H. et al. Stacked predictive sparse decomposition for classification of histology sections. Int. J. Comput. Vis. 113, 3–18 (2015).
Anand, V., Gupta, S., Koundal, D. & Singh, K. Fusion of U-Net and CNN model for segmentation and classification of skin lesion from dermoscopy images. Expert Syst. Appl. 213, 119230 (2023).
Guleria, K., Sharma, S., Kumar, S. & Tiwari, S. Early prediction of hypothyroidism and multiclass classification using predictive machine learning and deep learning. Measurement: Sens. 24, 100482 (2022).
Li, H., Pan, Y., Zhao, J. & Zhang, L. Skin disease diagnosis with deep learning: A review. Neurocomputing 464, 364–393 (2021).
Orhan, H. & Yavşan, E. Artificial intelligence-assisted detection model for melanoma diagnosis using deep learning techniques. Math. Modelling Numer. Simul. Appl. 3(2), 159–169 (2023).
Qasim Gilani, S., Syed, T., Umair, M. & Marques, O. Skin Cancer classification using deep spiking neural network. J. Digit. Imaging 23, 1–1 (2023)
Tembhurne, J. V., Hebbar, N., Patil, H. Y. & Diwan, T. Skin cancer detection using ensemble of machine learning and deep learning techniques. Multimedia Tools Appl. 16, 1–24 (2023).
Singh, S. K., Abolghasemi, V. & Anisi, M. H. Fuzzy logic with deep learning for detection of skin cancer. Appl. Sci. 13(15), 8927 (2023).
Patil, H. Frontier machine learning techniques for melanoma skin cancer identification and categorization: a thorough review. Oral Oncol. Rep. 19, 100217 (2024).
Yee, J., Rosendahl, C. & Aoude, L. G. The role of artificial intelligence and convolutional neural networks in the management of melanoma: A clinical, pathological, and radiological perspective. Melanoma Res. 34(2), 96–104 (2024).
Bhatt, H., Shah, V., Shah, K., Shah, R. & Shah, M. State-of-the-art machine learning techniques for melanoma skin cancer detection and classification: A comprehensive review. Intell. Med. 3(03), 180–190 (2023).
Ali, M. S., Miah, M. S., Haque, J., Rahman, M. M. & Islam, M. K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 5, 100036 (2021).
Salem, C., Azar, D. & Tokajian, S. An image processing and genetic algorithm-based approach for the detection of melanoma in patients. Methods Inf. Med. 57 (01/02), 74–80 (2018).
Ilkin, S. et al. Bacterial colony optimization algorithm based SVM for malignant melanoma detection. Eng. Sci. Technol. Int. J. 24(5), 1059–1071 (2021).
Li, L. F. et al. Deep learning in skin disease image recognition: A review. IEEE Access 8, 208264–208280 (2020).
Hosny, K. M., Kassem, M. A. & Foaud, M. M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PloS One 14(5), e0217293 (2019).
Patil, R. & Bellary, S. Machine learning approach in melanoma cancer stage detection. J. King Saud University-Computer Inform. Sci. 34(6), 3285–3293 (2022).
Bardou, D., Zhang, K. & Ahmad, S. M. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access 6, 24680–24693 (2018).
Ahammed, M., Al Mamun, M. & Uddin, M. S. A machine learning approach for skin disease detection and classification using image segmentation. Healthc. Analytics 2, 100122 (2022).
Balaji, V. R. et al. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 163, 107922 (2020).
Yu, H. Q. & Reiff-Marganiec, S. Targeted ensemble machine classification approach for supporting IoT enabled skin disease detection. IEEE Access 9, 50244–50252 (2021).
Ahmad, B. et al. Discriminative feature learning for skin disease classification using deep convolutional neural network. IEEE Access 8, 39025–39033 (2020).
Srinivasu, P. N. et al. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 21(8), 2852 (2021).
Shanthi, T., Sabeenian, R. S. & Anand, R. Automatic diagnosis of skin diseases using convolution neural network. Microprocess. Microsyst. 76, 103074 (2020).
Bajwa, M. N. et al. Computer-aided diagnosis of skin diseases using deep neural networks. Appl. Sci. 10(7), 2488 (2020).
Cai, G. et al. A multimodal transformer to fuse images and metadata for skin disease classification. Visual Comput. 39(7), 2781–2793 (2023).
Naeem, A. et al. SNC_Net: Skin Cancer detection by integrating handcrafted and deep learning-based features using Dermoscopy images. Mathematics 12(7), 1030 (2024).
Naeem, A., Anees, T. & DVFNet A deep feature fusion-based model for the multiclassification of skin cancer utilizing dermoscopy images. Plos One 19(3), e0297667 (2024).
Riaz, S., Naeem, A., Malik, H., Naqvi, R. A. & Loh, W. K. Federated and transfer learning methods for the classification of Melanoma and Nonmelanoma skin cancers: A prospective study. Sensors 23(20), 8457 (2023).
Naeem, A., Anees, T., Fiza, M., Naqvi, R. A. & Lee, S. W. SCDNet: A deep learning-based framework for the multiclassification of skin cancer using dermoscopy images. Sensors 22(15), 5652 (2022).
Naeem, A. & Anees, T. A multiclassification framework for skin cancer detection by the concatenation of Xception and ResNet101. J. Comput. Biomedical Inf. 6(02), 205–227 (2024).
Naeem, A., Khan, A. H., u din Ayubi, S. & Malik, H. Predicting the metastasis ability of prostate cancer using machine learning classifiers. J. Comput. Biomedical Inf. 4(02), 1–7 (2023).
Ayesha, H., Naeem, A., Khan, A. H., Abid, K. & Aslam, N. Multi-classification of skin cancer using multi-model fusion technique. J. Comput. Biomedical Inf. 5(02), 195–219 (2023).
Goceri, E. Diagnosis of skin diseases in the era of deep learning and mobile technology. Comput. Biol. Med. 134, 104458 (2021).
Rao, G. M., Ramesh, D., Gantela, P. & Srinivas, K. A hybrid deep learning strategy for image based automated prognosis of skin disease. Soft. Comput. 23, 1–2 (2023r).
Khan, M. A., Muhammad, K., Sharif, M., Akram, T. & Kadry, S. Intelligent fusion-assisted skin lesion localization and classification for smart healthcare. Neural Comput. Appl. 36(1), 37–52 (2024).
Aggarwal, M. et al. Privacy preserved collaborative transfer learning model with heterogeneous distributed data for brain tumor classification. Int. J. Imaging Syst. Technol. 34(2), e22994 (2024).
Lohith, R., Govinda, N. N., Pruthvi, K., Janhavi, V. & Gururaj, H. L. Facial skin disease detection using image processing. Int. J. Bioinf. Intell. Comput. 2(1), 1–1 (2023).
Cervantes, J., Garcia-Lamont, F., Rodríguez-Mazahua, L. & Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 408, 189–215 (2020).
Elsonbaty, A., Alharbi, M., El-Mesady, A. & Adel, W. Dynamical analysis of a novel discrete fractional lumpy skin disease model. Partial Differ. Equations Appl. Math. 9, 100604 (2024).
Kaur, P. et al. A hybrid convolutional neural network model for diagnosis of COVID-19 using chest X-ray images. Int. J. Environ. Res. Public Health 18(22), 12191 (2021).
Mishra, A. M., Kaur, P., Singh, M. P. & Singh, S. P. A self-supervised overlapped multiple weed and crop leaf segmentation approach under complex light condition. Multimedia Tools Appl. 30, 1–26 (2024) .
Kalpana, B., Reshmy, A. K., Pandi, S. S. & Dhanasekaran, S. OESV-KRF: optimal ensemble support vector kernel random forest based early detection and classification of skin diseases. Biomed. Signal Process. Control 85, 104779 (2023).
Moturi, D., Surapaneni, R. K. & Avanigadda, V. S. Developing an efficient method for melanoma detection using CNN techniques. J. Egypt. Natl Cancer Inst. 36(1), 6 (2024).
Natha, P. & Rajeswari, P. R. Skin cancer detection using machine learning classification models. Int. J. Intell. Syst. Appl. Eng. 12(6s), 139–145 (2024).
Kaur, P. et al. DELM: Deep ensemble learning model for multiclass classification of super-resolution leaf disease images. Turkish J. Agric. Forestry 47(5), 727–745 (2023).
Mijwil, M. M. Skin cancer disease images classification using deep learning solutions. Multimedia Tools Appl. 80(17), 26255–26271 (2021).
Kalouche, S., Ng, A. & Duchi, J. Vision-based classification of skin cancer using deep learning. conducted on Stanfords Machine Learning course (CS 229) taught. (2015).
Hossin, M. A. et al. Melanoma skin cancer detection using deep learning and advanced regularizer. In 2020 International Conference on Advanced Computer Science and Information Systems (ICACSIS) pp. 89–94. (IEEE, 2020).
Vijayalakshmi, M. M. Melanoma skin cancer detection using image processing and machine learning. Int. J. Trend Sci. Res. Dev. (IJTSRD) 3(4), 780–784 (2019).
Agrahari, P., Agrawal, A. & Subhashini, N. Skin cancer detection using deep learning. In Futuristic Communication and Network Technologies: Select Proceedings of VICFCNT 2020 (pp. 179–190. (Springer Singapore, 2022).
Gouda, W., Sama, N. U., Al-Waakid, G., Humayun, M. & Jhanjhi, N. Z. Detection of skin cancer based on skin lesion images using deep learning. In Healthcare, Vol. 10(7) p. 1183. (MDPI, 2022).
Funding
No funding has been received for this research or manuscript.
Author information
Authors and Affiliations
Contributions
V.K., P.K., S.G. and A.M.M.: write original draft; P.S. and M.D.: writing, review and editing; A.B. and I.G.: validation and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khullar, V., Kaur, P., Gargrish, S. et al. Minimal sourced and lightweight federated transfer learning models for skin cancer detection. Sci Rep 15, 2605 (2025). https://doi.org/10.1038/s41598-024-82402-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82402-x
Keywords
This article is cited by
-
Advancements and challenges of federated learning in medical imaging: a systematic literature review
Artificial Intelligence Review (2026)