Abstract
To evaluate the diagnostic value of metagenomic next-generation sequencing (mNGS) and galactomannan (GM) testing in invasive pulmonary aspergillosis (IPA) and to compare mNGS with other diagnostic approaches (serum/bronchoalveolar lavage fluid (BALF)-GM and conventional microbiological tests (CMTs) including sputum smears and culture, BALF fungal culture, and bronchial brushing). In all, 237 patients were enrolled in this retrospective study, including 120 patients with IPA and 117 with non-IPA pulmonary infections treated at Henan Provincial People’s Hospital between June 2021 and February 2024. The diagnostic performance of mNGS was compared to conventional diagnostic methods including serum GM, BALF-GM, sputum smear microscopy, sputum culture, bronchial brushings, and BALF culture. The proportion of patients with underlying diseases was significantly higher in the IPA group than in the non-IPA group (P < 0.05). Compared to conventional diagnostic methods for IPA, mNGS showed higher diagnostic efficacy, with a sensitivity of 92.5% and a specificity of 94.02%. The area under the receiver operating characteristic curve (AUC) for BALF-GM for diagnosing IPA was 0.8, with an optimal cutoff value of 0.546, sensitivity of 66.7%, and specificity of 82.1%. The combination of mNGS and BALF-GM testing further improved diagnostic performance (sensitivity of 96.67% and specificity of 78.63%). mNGS testing has excellent diagnostic efficacy for IPA, which is further enhanced by combining it with BALF-GM testing. This approach has considerable potential for the early diagnosis and targeted treatment of IPA.
Similar content being viewed by others
Introduction
Invasive pulmonary aspergillosis (IPA) is the most severe form of Aspergillus-related pulmonary infection and has a poor prognosis and a high mortality rate1. IPA primarily affects immunocompromised individuals, such as those with hematologic diseases, recipients of organ transplants, patients with cancer undergoing chemotherapy or radiation, and those with neutropenia2,3. It also occurs in patients with chronic obstructive pulmonary disease (COPD), bronchiectasis, diabetes, pulmonary tuberculosis, and coronavirus disease 2019 (COVID-19)4,5,6. Its clinical presentation is nonspecific, which complicates the diagnostic process. This issue is particularly pronounced in non-neutropenic IPA, which tends to progress rapidly and has a worse prognosis7,8. Hence, early and accurate diagnosis of IPA is important.
Although the gold standard for diagnosis of IPA is the detection of Aspergillus via histopathological examination and culture of clinical specimens, the sensitivity of fungal smears and the delayed results from fungal cultures hamper its early detection. Furthermore, patients with suspected IPA frequently have underlying pulmonary structural issues, severe thrombocytopenia, and other critical conditions, which increase the risk of histopathological examination by computed tomography (CT)-guided percutaneous or transbronchoscopic biopsy. Testing for galactomannan (GM) in serum and bronchoalveolar lavage fluid (BALF) is commonly used to diagnose aspergillosis. However, it is susceptible to interference from antifungal treatments and neutrophil counts, which can lead to false negatives. Furthermore, there is no agreement on an appropriate threshold, so manufacturers’ analytical thresholds are used2,9,10. Consequently, there is a pressing need to identify noninvasive or minimally invasive markers that enable the rapid and accurate diagnosis of IPA.
Metagenomic next-generation sequencing (mNGS) is a non-targeted, high-throughput, and sensitive nucleic acid-based technique used to detect pathogens causing infections of the lower respiratory tract11. It has shown promise for diagnosing infectious diseases but its use for IPA is supported mainly by case reports and small studies12,13,14. Further studies are needed to validate these results. Here, we investigated the value of mNGS and GM testing for the early diagnosis of IPA compared to conventional microbiological tests (CMTs).
Materials and methods
Study design and participants
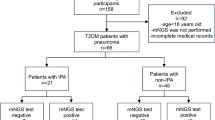
The protocols were approved by the Medical Ethics Committee of Henan Provincial People’s Hospital (approval no. 2023151) and were performed in accordance with its regulations and guidelines. This retrospective study involved 120 patients clinically diagnosed with IPA at Henan Provincial People’s Hospital between June 2021 and February 2024. The control group included 117 patients with non-IPA pulmonary infections. All of the enrolled patients had undergone mNGS testing. According to the criteria of the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium (EORTC/MSGERC)15, the diagnosis of IPA is based on the following: host risk factors for Aspergillus infection; clinical features, including persistent fever unresponsive to antibiotics for 96 h, and typical imaging characteristics such as patchy infiltrates with or without a halo sign, air-crescent sign, cavitation, multiple nodules, and wedge-shaped and segmental or lobar consolidation; mycological evidence, including microscopical detection of fungal elements in valid respiratory tract specimens, positive Aspergillus polymerase chain reaction (PCR), and any single serum GM ≥ 1.0 or BALF-GM ≥ 1.0 or single serum GM ≥ 0.7 and BALF-GM ≥ 0.8. According to these criteria, IPA is categorized as proven IPA (based on histopathological examination and/or culture positivity for Aspergillus), probable IPA (based on clinical diagnosis, host factors, clinical features, and at least one mycological criterion), and possible IPA (based on host factors and clinical features but without mycological evidence). Of the 120 patients with IPA enrolled in this study, which we termed clinically diagnosed IPA, 62 had proven IPA and 58 had probable IPA.
Collection of clinical data
Clinical data were collected from the electronic medical records system, including variables such as sex, age, hospitalization duration, underlying diseases, clinical symptoms, laboratory-related parameters, imaging features, histopathological examination results, treatments, and outcomes.
Conventional microbiological tests
All participants underwent bronchoscopy conducted by an experienced endoscopist according to the 2019 Guideline for Diagnostic Flexible Bronchoscopy in Adults16. Based on CT of the chest, the most affected segmental or subsegmental bronchi were selected for bronchoalveolar lavage and bronchial brushing to obtain BALF and bronchial brush specimens. This method can reduce specimen contamination by directly obtaining specimens from the lower respiratory tract. Extracted BALF was placed in a sterile container and immediately transported to the microbiology laboratory for mNGS testing and CMTs. Blood samples and suitable sputum samples were also collected.
Respiratory specimens (sputum, BALF, and bronchial brushings) were subjected to fluorescence microscopy and fungal fluorescence staining (Jiangsu Lifetime Biological Technology Co., Ltd., China) to visualize filamentous fungi. A sample was considered positive if septate hyaline hyphae branching at 45 degrees, suggesting Aspergillus species, were observed.
Sputum and BALF were cultured using Sabouraud dextrose agar (SDA) at 25 °C for 5 days. Fluffy colonies were transferred to potato dextrose agar (PDA) and incubated at 25℃ for 3–7 days for evaluation of colony characteristics and conidiophore morphology. Aspergillus species were identified via matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Bruker, Germany) and/or morphology.
BALF supernatant samples and serum were analyzed for GM using a Dynamiker Aspergillus Galactomannan Assay (Dynamiker Biotechnology [Tianjin] Co., Ltd., China), which is based on a sandwich enzyme-linked immunosorbent assay (ELISA). Samples were evaluated using the DNK-A600 Automated ELISA Reader (Dynamiker Biotechnology) according to the manufacturer’s recommendations. A serum GM > 0.5 ng/mL and BALF-GM > 0.8 ng/mL were considered positive according to the threshold stipulated in the manufacturer’s instructions.
mNGS procedures
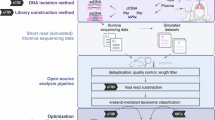
Host depletion, DNA extraction, library construction and sequencing
Lower respiratory tract infections (LRTIs) were diagnosed using a DNA-based mNGS standard operating procedure. Briefly, pathogens and human cells were separated from 1 mL BALF samples via centrifugation at 12,000 g for 5 min. Host nucleic acid was removed from the precipitate using 1 U Benzonase (Sigma) and 0.5% Tween 20 (Sigma) at 37 °C for 5 min. Terminal buffer (400 µL) was added to halt the reaction. The mixture (600 µL) was transferred to a new tube containing 500 µL ceramic beads and homogenized using Minilys Personal TGrinder H24 Homogenizer (Tiangen, China). Nucleic acids were extracted and eluted from 400 µL pretreated samples using a QIAamp UCP Pathogen Mini Kit in 60 µL elution buffer (Qiagen, Germany). DNA was quantified using a Qubit dsDNA HS Assay Kit (Invitrogen, CA)17,18. Next, cDNA was produced from 10 µL purified RNA. The KAPA Low-Throughput Library Construction Kit (KAPA Biosystems) was used to create a DNA/cDNA library in accordance with the manufacturer’s instructions. Library quality was assessed using the Qubit dsDNA HS Assay Kit and High-Sensitivity DNA Kit (Agilent) on an Agilent 2100 Bioanalyzer. Library pools were loaded onto an Illumina Nextseq CN500 sequencer for 75 cycles of single-end sequencing, generating approximately 20 million reads per library.
Bioinformatics analysis
Trimmomatic software was used to eliminate low-quality reads, duplicate reads, adapter contamination, and reads shorter than 70 bp19. Low-complexity reads were removed using Kcomplexity with the default settings. Human sequence data were located and eliminated using SNAP v. 1.0beta.18 to match the human sequence data to the hg38 reference genome20. The Kraken 2 criteria for selecting representative assemblies for microorganisms (bacteria, viruses, fungi, protozoa, and other multicellular eukaryotic pathogens) from the NCBI Assembly and Genome databases (https://benlangmead.github.io/aws-indexes/k2) were used to create the microbial genome database. Microbial reads were aligned to the database using Burrows-Wheeler Aligner software21. Reads with 90% identity to the reference were defined as mapped reads. In addition, reads with multiple locus alignments within the same genus were excluded from the secondary analysis. Only reads mapped to the genome of the same species were considered. The clinical reportable ranges (CRRs) for pathogens were established according to the Johns Hopkins ABX Guide22 (https://www.hopkinsguides.com/hopkins/index/Johns_Hopkins_ABX_Guide/Pathogens), The Manual of Clinical Microbiology23, and clinical case reports or research articles published in peer-reviewed journals.
Criteria for a positive mNGS result
The specifically mapped read number (SMRN) of each microbial taxon was normalized to SMRN per 20 million (M) of total sequencing reads (standardized SMRN, SDRN). The criteria for reporting mNGS results were as follows24,25: SDRN ≥ 3 (mycobacteria excluded) for bacteria, SDRN ≥ 3 for fungi/DNA viruses, SDRN ≥ 1 for RNA viruses, SDRN ≥ 100 for parasites, SDRN ≥ 3 for Mycoplasma/Chlamydia species, SDRN ≥ 1 for the Mycobacterium tuberculosis (MTB) complex, and positive mNGS for Nocardia species.
Quality control
To monitor sources of potential contamination, NC and sterile deionized water as non-template controls were prepared in parallel in each batch24. We used sterile cotton swabs dipped in sterile deionized water to wipe the surfaces of the centrifuge and biosafety cabinet to generate a list of the background microorganisms present in our laboratory.
Statistical analysis
Statistical analyses were performed using R (v. 4.3.3). pROC (v. 1.18.5) was used to calculate the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), Youden Index, and area under the receiver operating characteristic curve (AUC). ggplot2 (v. 3.5.1) was used to draw ROC curves. Normally distributed continuous data are expressed as means ± standard deviations, and between-group comparisons were performed using independent ttests. Non-normally distributed continuous data are expressed as medians (interquartile ranges) and were compared using Mann–Whitney Utests. Categorical data are described as frequencies and percentages and were compared using χ2 or Fisher’s exact tests. Clinical diagnosis of IPA was used as the gold standard to calculate sensitivity, specificity, PPV, and NPV. The diagnostic performances of serum GM and BALF-GM were assessed using three reported cutoff values1,7,9, and ROC curves were plotted. The optimal cutoff value for BALF-GM was determined using the Youden Index, and the use of mNGS and BALF-GM testing in combination for IPA was evaluated via serial and parallel analyses. The serial analysis was defined as both mNGS and BALF-GM being positive for Aspergillus, while the parallel analysis was defined as either mNGS or BALF-GM being positive for Aspergillus. Statistical significance was set as a two-sided Pvalue < 0.05.
Results
Clinical characteristics
Among the 237 patients, there were 120 IPA and 117 non-IPA patients. There were no statistically significant differences in sex or age between the groups (P > 0.05). The proportion of patients with underlying diseases was significantly higher in the IPA group than the non-IPA group. The CT imaging findings differed between the groups (Table 1).
Diagnostic performance of mNGS and conventional microbiological methods
Using clinical diagnosis of IPA as the reference standard, mNGS identified 111 Aspergillus-specific sequences in 120 patients with IPA and 7 in 117 patients who did not have IPA. The mNGS and CMTs showed high consistency (89 of 120, 74.2%) for diagnosing IPA, whereas mNGS identified the only mycological evidence in 31 IPA cases that were missed by CMTs. In comparison, nine patients with IPA missed by mNGS were diagnosed by CMTs. The sensitivity, specificity, PPV, and NPV of mNGS were 92.5%, 94.02%, 94.07%, and 92.44%, respectively. The sensitivity of a BALF GM ≥ 1 for diagnosing IPA was 48.33% with a specificity of 94.02%, which was second only to mNGS. Other conventional microbiological methods showed notably lower sensitivity than mNGS (Table 2).
Diagnostic performance of different cutoff values for serum GM and BALF-GM
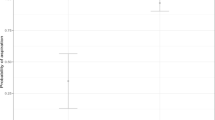
For serum GM tests, at cutoff values of 0.5, 0.7, and 1, sensitivity decreased to 35%, 27.5%, and 18.33%, respectively, and specificity increased to 93.16%, 96.58%, and 97.44% (Table 3). For BALF-GM tests, sensitivity decreased to 67.5%, 53.33%, and 48.33% with cutoff values of 0.5, 0.8, and 1, respectively, and specificity increased to 79.49%, 88. 89%, and 94.02% (Table 4). The AUC for serum GM detection was 0.61. The optimal cutoff value for serum GM, determined using the Youden Index, was 0.295, corresponding to a sensitivity of 46.7% and specificity of 85.5%. The AUC for BALF GM detection was 0.8. The optimal cutoff value for BALF-GM, determined using the Youden Index, was 0.546, corresponding to a sensitivity of 66.7% and specificity of 82.1% (Fig. 1).
Diagnostic performance of mNGS and BALF-GM testing in combination for IPA
Using a BALF-GM cutoff value of 0.546, the diagnostic performance of mNGS combined with BALF-GM for IPA was calculated using serial and parallel analyses; the results are given in Table 5; Fig. 2.
Discussion
As a group of pulmonary mycotic diseases, IPA often affects immunocompromised patients and can take an angio-invasive or airway-invasive form1,26. The incidence of IPA has increased in recent years because of the aging population; the widespread use of broad-spectrum antibiotics, antineoplastic drugs, and immunosuppressants; the COVID-19 pandemic; and the high frequency of invasive procedures and organ transplantation27,28,29,30. Given the rapid progression of IPA, early diagnosis is crucial to improve outcomes. However, its diagnosis is hampered by a lack of specific clinical symptoms and the difficulty obtaining tissue, enhancing the importance of microbiological evidence31,32,33.
Fungal culture of respiratory specimens is the most used diagnostic method34,35 but is hampered by contamination and airway colonization, and it is time-consuming with low diagnostic efficiency36,37. Direct smear microscopy, although more rapid than culture, depends on the microbial load in the sample and the skill of the microbiologist, resulting in low sensitivity and potential bias10,38. In this study, sputum smear, bronchial brushings, and BALF cultures had sensitivities < 30%, and sputum culture had the lowest sensitivity (13.33%). These limitations make these methods unsuitable for the early, rapid, and accurate diagnosis of IPA.
GM testing is an immunological method for the diagnosis of IPA. Serum GM testing has low sensitivity and is prone to false negatives, and it is useful primarily for diagnosing IPA in neutropenic patients39. In non-neutropenic patients, the sensitivity of BALF-GM testing is superior to serum GM testing15,40. However, BALF-GM testing can be affected by, for example, β-lactam antibiotics, intravenous drugs containing galactomannan, antifungal medications, mild disease, and low antigen concentrations, leading to false-positive and false-negative results. Moreover, the optimum cutoff values are unclear9,10. In this study, BALF-GM testing had a sensitivity of 48.33% for diagnosing IPA, significantly higher than serum GM (18.33%) and conventional respiratory-specimen methods, suggesting that BALF-GM has potential for the early diagnosis of IPA. The optimum cutoff for BALFGM is unclear, and the sensitivity of serum GM and BALF-GM is 24.3–46.9% and 54.5–80.0%, respectively, according to the cutoff value used41,42,43 In this study, we compared the diagnostic utility of several cutoff values for GM testing; sensitivity decreased as the cutoff value increased. For accurate diagnosis of IPA, the optimal cutoff value for BALF-GM, determined using an ROC curve and the maximum Youden Index, was 0.546, which had a sensitivity of 66.7% and specificity of 82.1%. A variety of factors influence GM results, so this BALF-GM cutoff value may be suitable at our institution but possibly not at others.
mNGS testing involves the extraction of microbial nucleic acids from clinical specimens followed by high-throughput sequencing and bioinformatics to identify microbes, thereby enabling the detection of the pathogens causing infectious diseases44. However, its diagnostic utility in IPA is unclear. Some studies have suggested that it is more rapid, sensitive, and precise than conventional microbiological methods, although those studies had small sample sizes and were retrospective45,46,47,48. We evaluated its diagnostic utility in a large number of IPA cases, comparing it sputum smear microscopy, sputum culture, BALF culture, and bronchial brushings, as well as serum and BALF-GM methods. We also explored the diagnostic utility of mNGS and BALF-GM testing in combination. mNGS alone had a sensitivity of 92.5% for diagnosing IPA, which increased to 96.67% when combined with BALF-GM testing, with a specificity of 78.63%. This performance significantly surpasses conventional microbiological methods. Given the rapid disease progression and poor prognosis of IPA, particularly in high-risk populations, there is a need for highly sensitive diagnostic methods to facilitate early antifungal treatment. When interpreting reports on the use of mNGS for IPA, the focus ought to be not only on the number of specific sequences and genome coverage of the identified pathogenic fungi but also the clinical manifestations, imaging findings, patient-specific factors, the results of other microbiological and pathological tests, and other indirect indicators of infection. The ability of mNGS to identify diverse pathogens is an important advantage. In patients with IPA, who have a high rate of mixed infections, mNGS facilitates the detection and differentiation of co-pathogens, thereby enabling selection of anti-infective treatments.
The use of mNGS in routine clinical practice has several issues. The need for costly and time-consuming (24–48 h) data analysis is a barrier to its widespread use, particularly in developing countries. The interpretation of mNGS results lacks universal consensus, hampering the determination of whether the microorganisms detected are pathogens or colonizers. Clinicians must consider the characteristics, laboratory tests, and imaging findings of suspected cases. Therefore, mNGS cannot replace conventional microbiological tests.
This study had several limitations. First, as a single-center, retrospective study, selection bias was inevitable. Second, Aspergillus PCR was not used to validate the diagnosis because this test is not routinely performed at our hospital. Finally, the clinical diagnostic standard used for case inclusion acknowledges that even experienced physicians cannot avoid clinical misjudgments, which can lead to discrepancies in diagnostic efficacy among methods. Therefore, the utility of mNGS for the diagnosis of IPA requires further clinical validation.
Conclusion
In this study, mNGS had good diagnostic performance for IPA, which was enhanced by its combination with BALF-GM testing. It enabled the identification of pathogens in mixed infections, underscoring its utility for the early diagnosis and targeted treatment of IPA.
Data availability
The datasets analyzed during the current study are available in the figshare dataset repository (https://doi.org/10.6084/m9.figshare.26942920.v1 ).
References
Denning, D. W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438 (2024).
Douglas, A. P. et al. Consensus guidelines for the diagnosis and management of invasive aspergillosis, 2021. Intern. Med. J. 51 (Suppl 7), 143–176 (2021).
von Lilienfeld-Toal, M. et al. Invasive Fungal Infect. Dtsch. Arztebl Int. 116, 271–278. (2019).
[Expert consensus on the. Diagnosis and treatment of pulmonary aspergillosis in patients with chronic obstructive pulmonary disease]. Zhonghua Jie He He Hu Xi Za Zhi. 47, 604–622 (2024).
Lai, C. C. & Yu, W. L. COVID-19 associated with pulmonary aspergillosis: a literature review. J. Microbiol. Immunol. Infect. 54, 46–53 (2021).
Zhuang, Q. et al. Galactomannan in Bronchoalveolar Lavage Fluid for Diagnosis of Invasive Pulmonary Aspergillosis with Nonneutropenic Patients. Can Respir J. 2017: 3685261. (2017).
Trof, R. J. et al. Management of invasive pulmonary aspergillosis in non-neutropenic critically ill patients. Intensive Care Med. 33, 1694–1703 (2007).
Park, S. Y. et al. Computed tomography findings in invasive pulmonary aspergillosis in non-neutropenic transplant recipients and neutropenic patients, and their prognostic value. J. Infect. 63, 447–456 (2011).
Gerard, R. et al. Is there still a place for serum galactomannan in the diagnosis of invasive aspergillosis in children at high risk and under antifungal prophylaxis? Mycoses. 67, e13764. (2024).
Park, S. Y., Ardura, M. I. & Zhang, S. X. Diagnostic limitations and challenges in current clinical guidelines and potential application of metagenomic sequencing to manage pulmonary invasive fungal infections in patients with haematological malignancies. Clin. Microbiol. Infect. 30, 1139–1146 (2024).
Chiu, C. Y. & Miller, S. A. Clinical metagenomics. Nat. Rev. Genet. 20, 341–355 (2019).
Yang, A. et al. Diagnosis by metagenomic next-generation sequencing of invasive pulmonary aspergillosis in an infant with chronic granulomatous disease. Respir Med. Case Rep. 41, 101792 (2023).
He, B. C. et al. The application of next-generation sequencing in diagnosing invasive pulmonary aspergillosis: three case reports. Am. J. Transl Res. 11, 2532–2539 (2019).
Miao, Q. et al. Microbiological Diagnostic performance of Metagenomic Next-generation sequencing when Applied to Clinical Practice. Clin. Infect. Dis. 67, S231–s240 (2018).
Donnelly, J. P. et al. Revision and update of the Consensus definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 71, 1367–1376 (2020).
[Guideline for diagnostic flexible bronchoscopy in adults. ]. Zhonghua Jie He He Hu Xi Za Zhi 2019; 42: 573–590. (2019).
Amar, Y. et al. Pre-digest of unprotected DNA by Benzonase improves the representation of living skin bacteria and efficiently depletes host DNA. Microbiome. 9, 123 (2021).
Zhou, Z. et al. Heightened Innate Immune responses in the respiratory tract of COVID-19 patients. Cell. Host Microbe. 27, 883–890e882 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014).
Zaharia, M. et al. Faster and more accurate sequence alignment with SNAP. https://arxiv.org/abs/1111.5572 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009).
Jing, C. et al. Clinical evaluation of an Improved Metagenomic Next-Generation sequencing test for the diagnosis of Bloodstream infections. Clin. Chem. 67, 1133–1143 (2021).
Versalovic, J. Manual of Clinical Microbiology (ASM, 2011).
Qin, H. et al. A retrospective paired comparison between untargeted next generation sequencing and Conventional Microbiology tests with wisely chosen metagenomic sequencing positive criteria. Front. Med. (Lausanne). 8, 686247 (2021).
Qian, Y. Y. et al. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front. Cell. Infect. Microbiol. 10, 567615 (2020).
Vergidis, P., Sendi, P., Alkhateeb, H. B. & Nguyen, M. H. How do I manage refractory invasive pulmonary aspergillosis. Clin. Microbiol. Infect. 30, 755–761 (2024).
Cabrera, N. L. et al. Novel agents in the treatment of invasive fungal infections in solid organ transplant recipients. Curr. Opin. Organ. Transpl. 27, 235–242 (2022).
Jia, H. et al. Diagnostic efficacy of metagenomic next generation sequencing in bronchoalveolar lavage fluid for proven invasive pulmonary aspergillosis. Front. Cell. Infect. Microbiol. 13, 1223576 (2023).
Muthu, V. et al. Risk factors, mortality, and predictors of survival in COVID-19-associated pulmonary mucormycosis: a multicentre retrospective study from India. Clin. Microbiol. Infect. 30, 368–374 (2024).
Lu, L. Y. et al. Prevalence, risk factors, clinical features, and Outcome of Influenza-Associated Pulmonary aspergillosis in critically ill patients: a systematic review and Meta-analysis. Chest. 165, 540–558 (2024).
Long, Z. et al. Improved diagnostic markers for invasive pulmonary aspergillosis in COPD patients. Front. Cell. Infect. Microbiol. 14, 1294971 (2024).
Kuo, C. W. et al. Navigating the challenges of invasive pulmonary aspergillosis in lung cancer treatment: a propensity score study. Ther. Adv. Med. Oncol. 15, 17588359231198454 (2023).
Tamkeviciute, L. et al. Different forms of pulmonary aspergillosis: a pictorial essay. Eur. J. Radiol. 171, 111290 (2024).
Matsumoto, K. et al. Molecular and ultrastructural morphological analyses of highly metamorphosed aspergillus fumigatus on human formalin-fixed paraffin-embedded tissue. Med. Mol. Morphol. 57, 326–332 (2024).
Niu, S., Liu, D., Yang, Y. & Zhao, L. Clinical utility of metagenomic next-generation sequencing in the diagnosis of invasive pulmonary aspergillosis in acute exacerbation of chronic obstructive pulmonary disease patients in the intensive care unit. Front. Cell. Infect. Microbiol. 14, 1397733 (2024).
Scharmann, U. et al. Microbiological non-culture-based methods for diagnosing invasive pulmonary aspergillosis in ICU patients. Diagnostics (Basel). 13, 2718 (2023).
Hardak, E. et al. Diagnostic role of polymerase chain reaction in bronchoalveolar lavage fluid for invasive pulmonary aspergillosis in immunocompromised patients - a retrospective cohort study. Int. J. Infect. Dis. 83, 20–25 (2019).
Cadena, J., Thompson, G. R. 3rd., & Patterson, T. F. Aspergillosis: epidemiology, diagnosis, and treatment. Infect. Dis. Clin. North. Am. 35, 415–434 (2021).
De Vlieger, G. et al. Beta-D-glucan detection as a diagnostic test for invasive aspergillosis in immunocompromised critically ill patients with symptoms of respiratory infection: an autopsy-based study. J. Clin. Microbiol. 49, 3783–3787 (2011).
Bupha-Intr, O. et al. Consensus guidelines for the diagnosis and management of invasive fungal disease due to moulds other than aspergillus in the haematology/oncology setting, 2021. Intern. Med. J. 51 (Suppl 7), 177–219 (2021).
Zhou, W. et al. Diagnostic value of Galactomannan Antigen Test in serum and Bronchoalveolar Lavage Fluid Samples from patients with nonneutropenic invasive pulmonary aspergillosis. J. Clin. Microbiol. 55, 2153–2161 (2017).
Dai, Z. et al. Comparing the diagnostic value of bronchoalveolar lavage fluid galactomannan, serum galactomannanan, and serum 1,3-β-d-glucan in non-neutropenic respiratory disease patients with invasive pulmonary aspergillosis. Med. (Baltim). 100, e25233 (2021).
Ghazanfari, M. et al. Comparative analysis of galactomannan lateral flow assay, galactomannan enzyme immunoassay and BAL culture for diagnosis of COVID-19-associated pulmonary aspergillosis. Mycoses. 65, 960–968 (2022).
Gu, W. et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124 (2021).
Shi, Y. et al. Metagenomic next-generation sequencing for detecting aspergillosis pneumonia in immunocompromised patients: a retrospective study. Front. Cell. Infect. Microbiol. 13, 1209724 (2023).
Zhu, N. et al. Performance of mNGS in bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in non-neutropenic patients. Front. Cell. Infect. Microbiol. 13, 1271853 (2023).
Bao, S. et al. Metagenomic next-generation sequencing for the diagnosis of pulmonary aspergillosis in non-neutropenic patients: a retrospective study. Front. Cell. Infect. Microbiol. 12, 925982 (2022).
Liu, Z. et al. Metagenomic next-generation sequencing for the diagnosis of invasive pulmonary aspergillosis in type 2 diabetes mellitus patients. Sci. Rep. 14, 16618 (2024).
Acknowledgements
Thank you Zhicheng Luo and Tiandong Li for your contribution to the data analysis in this article. Thank you for the technical support provided by the Vision Medicals Co. Ltd. (Guangzhou, China) in mNGS testing.
Funding
This study was supported by the Zhengzhou Collaborative Innovation Major Project (Zhengzhou University, 518-23240003/518-23240004).
Author information
Authors and Affiliations
Contributions
Each author had participated sufficiently in the work to take public responsibility for appropriate portions of the content. CCL, YY, and XYZ contributed to data collection. JY, HML and YXA designed this study. JY, QQZ, and XW performed the statistical analysis. JY and XW drafted the paper. All authors helped to interpret the data and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of Henan provincial People’s Hospital (Approve No: 2023151). All methods were carried out in accordance with the declaration of Helsinki. Written informed consent was obtained from all subjects.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, J., Wu, X., Zhang, Q. et al. Metagenomic next-generation sequencing and galactomannan testing for the diagnosis of invasive pulmonary aspergillosis. Sci Rep 14, 31389 (2024). https://doi.org/10.1038/s41598-024-82806-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82806-9