Abstract
Wnt signaling is a critical pathway implicated in cancer development, with Frizzled proteins, particularly FZD10, playing key roles in tumorigenesis and recurrence. This study focuses on the potential of repurposed FDA-approved drugs targeting FZD10 as a therapeutic strategy for nasopharyngeal carcinoma (NPC). The tertiary structure of human FZD10 was constructed using homology modeling, validated by Ramachandran plot and ProQ analysis. Virtual screening of 1,094 FDA-approved drugs identified 17 potential inhibitors, with prazosin, rilpivirine, doxazosin, and nicergoline demonstrating significant cytotoxicity against NPC cells. Further molecular dynamics simulations and binding energy analyses confirmed the stable binding of these drugs to FZD10. The results suggest that these repurposed drugs could serve as promising candidates for targeted NPC therapy, warranting further investigation.
Similar content being viewed by others
Introduction
Wnt signaling is a vital pathway involved in embryonic development and adult tissue regeneration1. It comprises two main pathways: the canonical and non-canonical Wnt pathways, both requiring Wnt ligands to bind to membrane receptor proteins to activate signal transduction. Interestingly, Wnt signaling has been extensively investigated in the development and progression of various cancers, such as gastric cancer, lung cancer, colorectal cancer, leukemia, bladder cancer, and nasopharyngeal cancer2,3,4,5,6. The Frizzled (FZD) protein family, which serves as membrane receptors in the Wnt signaling pathway, has implications for cancer development. For instance, FZD1 is diagnostically relevant in clear cell renal cell carcinoma7. FZD7 is upregulated in gastric and esophageal squamous cell carcinoma, facilitating β-catenin nuclear translocation, and activation of the WNT/β-catenin signaling pathway, including the epithelial-mesenchymal transition (EMT) process8,9. Meanwhile, FZD8’s high expression is crucial for the proliferation and metastasis of renal cell carcinoma10. FZD10, a member of the Frizzled protein family, has gained attention for its role in cancer. Initially identified as upregulated in cervical and colon cancers, FZD10 is absent in healthy adult tissues but becomes upregulated in the late stages of colon tumor progression11,12. Our previous transcriptomic analysis identified enhanced Wnt signaling in recurrent nasopharyngeal carcinoma (NPC) tissues, with FZD10 significantly upregulated and correlated with NPC recurrence13. This discovery led to our focus on targeting FZD10 for NPC therapy. Inhibiting the Wnt signaling pathway is a key strategy for cancer therapy, with several FZD-targeted therapeutic drugs in development. For example, OMP18R5, a monoclonal antibody that binds to and blocks FZD function, has shown efficacy in inhibiting tumor growth14. Additionally, actinium-225-labeled OTSA101, a radioactive monoclonal antibody targeting FZD10, holds therapeutic potential for patients with synovial sarcoma15.
This study focuses on targeting FZD10 for cancer treatment, utilizing NPC as a model. NPC, a type of head and neck cancer originating in the nasopharynx, shows elevated incidence rates in specific ethnic and geographic populations, such as the Cantonese population in southern China and certain areas of Asia, the Arctic, and the Middle East/North Africa16. Due to its central location within the head, surgical excision of NPC tumors presents significant challenges. While radiation therapy is effective in early-stage NPC cases, advanced diagnosis often limits its efficacy, necessitating concurrent chemotherapy, frequently employing cisplatin in synergy with 5-fluorouracil17,18. Considering chemotherapy’s adverse effects, there is growing interest in targeted therapies that offer heightened specificity to the cancer while minimizing toxicity.
In recent years, computational approaches have emerged as critical tools in the early stages of drug discovery, shedding light on rapid screening and identification of hit compounds, along with guiding hit-to-lead optimization. Importantly, we can simulate and predict interactions between drug candidates and their biological targets in various aspects, e.g., thermodynamic and biochemical, using validated algorithms and molecular modeling techniques. These advancements could provide a thorough understanding of how molecules affect biological targets and modulate signaling cascades. For instance, researchers employed docking-based virtual screening and molecular dynamics simulations to conduct drug repurposing and to search for novel anti-cancer19,20,21,22,23,24, anti-inflammation25,26, anti-SARS-CoV-2 main protease27,28,29 agents, compound supplements for cultured meat production30, etc. Moreover, molecular docking has a variety of valuable applications. For example, the identification of Kamiya-8 as a potential sweetener with minimal binding to bitter receptors is particularly notable. In addition, the examination of SNPs in the centrosomal protein of 63 kDa (CEP63) gene demonstrates how in silico modeling can reveal the structural effects of genetic variations. A comprehensive analysis of mutations—such as the L61P variant and mutations in the tyrosinase protein associated with albinism—further highlights the utility of this approach for understanding genetic disorders31,32,33,34,35. Therefore, it is evident that integration of computational advancements with in vitro studies is advantageous not only for identifying promising agents, but also for providing a deeper understanding of protein-ligand structural dynamics, recognition, and susceptibility at the atomic level.
Targeting FZD10 for cancer therapy presents a promising avenue, particularly through the repurposing of existing drugs. Drug repurposing involves finding new therapeutic applications for FDA-approved medications, mitigating the risk of failure and reducing development costs36. Hence, our study aims to explore repurposed drugs for targeting FZD10 protein in NPC treatment.
Materials and methods
FZD10 protein construction
The tertiary structure of human FZD10 was generated using the intensive modeling mode of the Protein Homology/Analogy Recognition Engine V 2.0 (Phyre2) server, based on the homology model method37. The amino acid sequence of human FZD10, obtained from UniProt (https://www.uniprot.org/uniprot/Q9ULW2), was submitted to the Phyre2 server. Model validation was conducted using the RAMPAGE server (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). The RAMPAGE server checks the backbone conformation qualities of the model peptide based on the phi/psi distribution and displays the number of residues in favored, allowed, and outer areas in a Ramachandran plot. The Ramachandran plot was generated to assess the stereochemical quality of the model. More than 85% of residues fell within the favored and allowed regions, with minimal outliers, indicating that the backbone conformations are within acceptable limits for homology models. Furthermore, the ProQ (Protein Quality Prediction) server (https://proq.bioinfo.se/cgi-bin/ProQ/ProQ.cgi) was used to verify the quality of the FZD10 model. The ProQ is a neural network-based tool for assessing protein model quality. It evaluates models using two different measures: LGscore and MaxSub. Both scores detect segments in common between the predicted model and the correct target structure, serving as structural comparison scores (Sstr). LGscore employs a statistical distribution to relate the Sstr score to the probability of obtaining a higher score by chance (p-value), whereas MaxSub normalizes the Sstr by dividing it by the length of the correct target structure38. The binding pocket of FZD10 for repurposing drugs was identified prior to molecular docking. The complexed structure of FZD10 with the reference ligand (taladegib) model was generated using Patchdock, an online molecular docking algorithm39.
Structure-based virtual screening
A total of 1,094 small molecules, FDA-approved drugs with molecular weights between 300 and 600 g/mol, were retrieved from the DrugBank database (https://www.drugbank.ca). The canonical SMILES format of compound structures was downloaded from the PubChem chemistry database (https://pubchem.ncbi.nlm.nih.gov). Subsequently, 3D compound structures were generated using CORINA classic (Molecular Networks GmbH, Germany, and Altamira, LLC, USA). The FZD10 structure was employed for screening FDA-approved drugs using three protein-docking software: iGemdock v2.1 with the following settings: a population size of 200, 70 generations, and 2 solution numbers, FlexX software (LeadIT 2.3.2), and cDocker v3.0. These three computational tools were used for molecular docking and virtual screening. iGEMDOCK, a molecular docking and screening program, employs the GEMDOCK (Generic Evolutionary Method for molecular DOCKing) algorithm to predict the binding between small molecules (ligands) and target proteins (receptors) by optimizing binding conformations and interaction energies40. This program was used for initial small-molecule drug screening. FlexX is a fragment-based docking tool that breaks down ligands into fragments, docks each fragment into the active site of the target protein, and then incrementally reassembles them to create the final docking pose41. It offers high-speed docking, enabling large-scale screenings. cDocker, on the other hand, uses the CHARMM (Chemistry at HARvard Macromolecular Mechanics) force field and molecular dynamics to simulate ligand-protein interactions, providing highly accurate docking results. Integrating these three programs, the docking study gains a balanced approach that harnesses the unique strengths of each tool, leading to a thorough screening analysis of small molecular drugs targeting the FZD10 protein.
Effects of candidate FDA-approved drugs on NPC cells
A total of 3,000 NPC cells were plated in each well of flat-bottom 96-well tissue culture plates. After 24 h, the culture media (200 µL/well) was replaced with fresh medium supplemented with various screening concentrations (0.1, 1, 10, and 100 µM) of drugs, including prazosin, rilpivirine, doxazosin, and nicergoline. These logarithmic concentrations provide the broad range that encompasses the NPC cytotoxic doses of the compound. The positive control consisted of treatment with cisplatin. Following 48 h of treatment, cell viability was assessed using MTT assay. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) is a tetrazolium compound that is converted into a purple formazan product by metabolically active, viable cells. A stock solution of 5 mg/mL MTT was diluted in RPMI medium to a final concentration of 0.5 mg/mL and added to the plate. The cells were then incubated at 37 °C for 3 h. After incubation, the MTT solution was removed, and the formazan crystals were dissolved in DMSO. The absorbance of the resulting solution was measured at 540 nm to determine the results. The MTT assay was performed in triplicate, and statistical significance was assessed using a t-test, with p-values less than 0.05 considered significant. Furthermore, NPC colony formation was determined after 1–2 weeks of growth. Briefly, two hundred NPC cells were seeded in a 24-well tissue culture plate and incubated for 24 h. The growth medium was replaced, and further incubation occurred for 10 days (SUNE5-8 F) and 7 days (SUNE6-10B). The culture medium was replaced every 3 days. The surviving colonies were fixed with 4% paraformaldehyde for 10 min, washed three times with PBS, and stained with 0.5% crystal violet for 15 min. The plates were washed with tap water three times and dried overnight. The colonies were then documented by capturing images. To quantify the colony formation ability, ColonyArea42 was used to measure the intensity of the colonies after staining with crystal violet.
Western immunoblotting
To assess the specificity of repurposed drugs on the FZD10 protein, we examined the expression of Wnt signaling proteins in SUNE5-8 F and SUNE6-10B cells. These cells were cultured in complete RPMI medium supplemented with IC25 and IC50 concentrations of drugs for 48 h. Subsequently, whole-cell lysate protein was analyzed to determine the levels of Wnt signaling protein expression, including non-phosphorylated β-catenin and phosphorylated LRP6 (P-LRP6) through Western blotting. Briefly, cells were scraped from 10 cm culture dishes. The pellets were washed three times with PBS at pH 7.4 and then lysed using a lysis buffer (50 mM Tris-HCl, pH 7.0, 5 mM EDTA, 150 mM NaCl, 5 mM DTT, and 1% IGEPAL) for 10 min on ice. After centrifugation at 12,000 × g for 10 min at 4 °C, protein concentrations were determined using the Bradford protein assay (Bio-Rad). Twenty micrograms of proteins were subjected to 10% SDS-PAGE (PageRuler Plus Prestained Protein Ladder, Thermo Scientific), which were then transferred onto a nitrocellulose membrane. The membrane was then incubated in a 5% bovine serum albumin (BSA) for 1 h and probed with primary antibodies overnight at 4 °C. The primary antibodies used included anti-FZD10 (#ab83044; Abcam, USA), anti-non-phosphorylated β-catenin (#19807; Cell Signaling Technology), and anti-P-LRP6 (#2568; Cell Signaling Technology). After washing away unbound antibodies, the membrane was incubated with HRP-conjugated secondary antibodies (Cell Signaling Technology) at room temperature for 1 h. The membrane was washed three times with TBST. The target proteins were detected using an ECL reagent (BIO-RAD) and visualized with a chemiluminescent Western blot imaging system (G: BOX, SYNGENE).
Immunofluorescence
Initially, 8,500 cells were seeded into 8-well chamber glass slides (Nunc™ Lab-Tek™), and after 24 h, IC50 concentrations of drugs were added to each well. A control group with 0.5% DMSO was also included. After 48 h, the adherent cells were fixed with 4% paraformaldehyde, washed, and permeabilized with 0.25% Triton-X-100 for 10 min at 4 °C. Subsequently, the cells were blocked in 3% BSA for 1 h and incubated with a 1:200 dilution of the primary antibody anti-non-phosphorylated β-catenin (#19807; Cell Signaling Technology) overnight. After washing, the cells were incubated with a 1:200 fluorochrome-conjugated secondary antibody, anti-rabbit (#4414s; IgG (H + L), F(ab’)2 fragment (Alexa Fluor® 647 Conjugate)) for 1 h and washed four times for 5 min each. The actin filament was stained by incubating with a 1:500 dilution of for 30 min. The nucleus was stained with 5 µg/mL Hoechst 33,342 for 2 min. The stained cells were analyzed using a confocal fluorescence microscope (Fluoview FV10i, Olympus). The quantitative fluorescent value was calculated using the corrected total cell fluorescence (CTCF) formula, where CTCF = Integrated Density - (Area of selected cell × Mean fluorescence of background readings).
Nuclear magnetic resonance
Saturation transfer difference (STD) Nuclear Magnetic Resonance Spectroscopy (STDNMR) spectra were acquired on a Bruker Avance Ascend 600 MHz Spectrometer (Bruker, Switzerland) equipped with a 5 mm BBO Prodigy CryoProbe. The temperature of the probe was set to 298 K. Then, 230 nM protein and 0.3–1.5 µM ligand was dissolved in 600 µl of deuterium oxide (D2O) into a 5 mm NMR tube. The central solvent (D2O) peak was used as an internal chemical shift reference point (δH 4.70). The spectrometer frequency was set at 600 MHz for 1 H and STD dimension detection. Spectral widths of 1 H dimensions were 9600 Hz with a 30◦ pulse angle. The number of complex points collected for the 1 H dimension was 8 with a recycle delay of 2.5 s and 8 scans.
Molecular dynamics simulations
The initial structures of FZD10 and drug candidates were obtained from the docking pose predicted by the cDocker docking program. The protonation states of ionizable amino acids were predicted at physiological pH (pH = 7.4) using the PDB2PQR web interface (https://server.poissonboltzmann.org/) while partial atomic charges for drug candidates were approximated by the AMBER force field version 2 (GAFF2) within the antechamber module implemented in the AMBER22 software package. The constructed complexes were then simulated under periodic boundary conditions with the isothermal-isobaric (NPT) scheme (310 K, 1 atm). The AMBER ff14SB force field and the generalized AMBER force field version 2 (GAFF2) were used to treat bonded and non-bonded interactions43, while solvents were introduced using the TIP3P water model. The overall system charge was neutralized by randomly adding either sodium or chloride ions44. Hydrogen atoms and water molecules were minimized using 500 steps of steepest descent (SD) followed by 1,500 steps of conjugated gradient (CG) methods, while the remaining molecules were fixed. Subsequently, the entire complex underwent minimization using the same procedure. The particle mesh Ewald summation and the SHAKE algorithm45 were utilized to manage electrostatic interactions and constrain all hydrogen atoms46. Temperature was increased from 10 to 310 K using a Langevin thermostat47 with a collision frequency of 2 ps− 1, while pressure was controlled by the Berendsen barostat48. The molecular dynamics (MD) production phase was conducted for 100 ns, increasing at 2 fs for each step. MD outputs were obtained from the cpptraj module, and the decomposition energy (\(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\)) was computed using Molecular Mechanics with Generalized Born and Surface Area Solvation (MM/GBSA) implemented in AMBER22. To estimate the ligand binding affinity, the end-point binding free energy (∆Gbind) of each complex was calculated based on the solvated interaction energy (SIE) approach35, where ∆Gbind is the sum of van der Waals (EvdW), electrostatic (Eele), reaction field (GRF), cavity (γΔSA(ρ)), and a constant (C) value. The coefficients were set to default values (α = 0.105, γ = 0.013, and C = -2.89).
Results and discussion
Construction of FZD10 protein structure
Before performing in silico screening, the 3D structure of FZD10 was constructed using homology modeling based on its amino-acid sequence available in Uniprot by using Phyre237. We found that 83% of FZD10’s residues have been successfully modeled with more than 90% confidence when compared to the Smoothened (SMO) membrane crystal structure co-crystallized with taladegib (PDB ID: 4JKV). The calculation relies on the quality of the alignment and the level of structural similarity to the templates. A confidence score exceeding 90% indicates that the predicted FZD10 structure is highly accurate in its modeled positions, likely reflecting the actual structure of the protein. With 83% of residues modeled at this high level of confidence, the FZD10 model is structurally robust for applications like molecular docking and virtual screening. FZD10 comprises two main parts including the N-terminal extracellular domain and the 7-helix transmembrane domain, as depicted in Fig. 1A. Additionally, a Ramachandran diagram was plotted to assess the quality of the predicted FZD10 structure. The constructed structure was considered “good quality” as it has 86.5% favored regions (Fig. 1B). Furthermore, the predicted secondary structure of FZD10 was evaluated for protein quality using a neural network-based protein quality predictor known as ProQ38. The predicted LGscore (negative logarithm of a structural P-value) was found to be 3.154, and the predicted MaxSub was 0.168, indicating a good quality model protein. To identify the predicted binding site, blind docking was performed by using the PATCHDOCK server, a geometry-based molecular docking algorithm39. This process guided the inhibitor binding region within FZD10 (Fig. 1A), which was then employed as a docking site for virtual screening.
Docking-based virtual screening
To rapidly screen potential compounds capable of inhibiting FZD10, we performed docking-based virtual screening of 1,094 FDA-approved drugs using three docking programs: FlexX, cDocker, and iGemdock. Candidates identified as exhibiting high binding affinity by multiple programs are likely to enhance the reliability of predictive outcomes. The interaction energy predicted by each program served as the primary parameter for ranking the drug candidates. Initially, we clustered the top 60 drugs with the lowest docking scores from iGemdock in the first-round screening, utilizing parameters as described in the materials and methods. These 60 compounds underwent further calculation of protein-ligand binding interactions using FlexX and cDocker, with a known drug, taladegib, serving as a reference (Fig. 2 and Table S1). For this phase, we exclusively selected non-oncology drugs, filtering them down to 47 compounds out of the initial 60. The docking results revealed eight compounds exhibiting lower interaction energy than the taladegib across all docking programs, including cefamandole, ceforanide, cefotaxime, cefotiam, doxazosin, imidurea, latamoxef, and paliperidone. To expand the number of screened candidates, nine additional compounds were considered from the remaining non-oncology drugs. Selection criteria included affordability and lower interaction energy in at least one docking program, compared to the reference compound. Subsequently, cefazolin, delamanid, methylergometrine, mirabegron, nicergoline, prazosin, rilpivirine, sparfloxacin, and valganciclovir were selected, resulting in a total of 17 drug candidates (marked with an asterisk in Fig. 2). The selected drug candidates, along with their interaction energy, were listed in Table 1.
In vitro repurposed drug screening in NPC cells
To further assess the potential therapeutic utility of the compounds for NPC, all 17 underwent toxicity testing via the MTT assay on NPC cells, revealing a concentration-dependent suppression of NPC cell growth within 48 h. In the preliminary NPC cytotoxicity screening, drugs with an IC50 greater than 100 µM for NPC cells were excluded, as such high doses are unlikely to effectively reduce NPC growth. The use of high drug concentrations may cause off-target or non-specific toxic effects, potentially harming healthy cells and tissues. Drugs requiring such high concentrations may lack a selective therapeutic window, posing toxicity risks to both cancerous and normal cells. Consequently, four compounds—prazosin, rilpivirine, doxazosin, and nicergoline—met the 50% inhibition cutoff at 100 µM and were quantified with a selectivity index (SI)49. The SI index represents the ratio of IC50 calculated for normal nasopharynx cell (NP460) and cancer cells, indicating a potential cytotoxic effect on NPC cells (Table 2). The IC50 values and SEM are detailed in Table S2. Consequently, the SI value for these four repurposed drugs is greater than 1, signifying a desirable level of selectivity towards NPC cells49. Furthermore, SUNE5-8 F and SUNE6-10B NPC cells exhibiting the highest selectivity index from the screening approach, were then used as a model of NPC to study the anticancer efficiency of the candidate compounds.
Molecular dynamics (MD) simulations
End-state binding energy predictions
To assess the compounds’ binding capability toward FZD10, the last 10-ns MD snapshots were utilized to estimate the binding energy (ΔGbind) based on the end-point Solvated Interaction Energy (SIE) approach. As listed in Table 3, the screened compounds exhibited ΔGbind values in the range of -9.83 to -7.76 kcal/mol, suggesting that all compounds could form stable complexes with FZD10 in the order of nicergoline > doxazosin > rilpivirine > prazosin, validated later by the STD-NMR. Particularly, van der Waals interaction energy (EVdW) was found to be the primary force stabilizing the protein-ligand complexes, approximately 4- to 15-fold lower than the coulombic interaction energy (Ecoul). Noticeably, prazosin, rilpivirine, and nicergoline showed a similar level of Ecoul (~ -3 to -4 kcal/mol), while doxazosin was more stabilized by Ecoul than other compounds (~ 4-fold lower of Ecoul). Furthermore, nicergoline exhibited the lowest solvation free energy (ΔGcavity) followed by doxazosin, rilpivirine, and prazosin, indicating that they could be well buried into the binding pocket.
System stability and structural dynamics
The stability of the modeled complexes was observed in terms of root-mean-square deviation (RMSD) within the 5 Å sphere of compounds, plotted along the simulated time of 100 ns. As shown in Fig. 3, the rilpivirine and nicergoline complex systems fluctuated less than the prazosin and doxazosin complexes. Particularly, the RMSD value of the nicergoline/FZD10 complex remained stable throughout the entire simulated time, while the rilpivirine/FZD10 complex gradually fluctuated after 80 ns but remained stable until 100 ns. In contrast, the prazosin/FZD10 and doxazosin/FZD10 complexes showed significant deviations at 50–70 ns compared to the other two complexes but became stable after 80 ns. The RMSD analysis suggests that all modeled complexes reach equilibrium after 80 ns and the snapshots at this state could be further analyzed. Additionally, nicergoline was observed to be well recognized by FZD10, resulting in a reduced magnitude of protein structural dynamics, supporting its highest stability to form a complex with FZD10. Note that the RMSD of the ligand and the backbone of the protein-ligand complex were also illustrated in Figure S1. Apart from RMSD analysis, we calculated the number of atoms surrounding each compound, which is a crucial parameter implying the ability of molecular recognition within the binding pocket. We found that the number of surrounding atoms averaged in the last 10 nanoseconds of each complex was in the order of doxazosin (339 atoms) > nicergoline (301 atoms) > rilpivirine (272 atoms) > prazosin (241 atoms) as shown in Fig. 3. The higher number of surrounding atoms of the doxazosin and nicergoline complexes may be the reason supporting the higher ΔGbind during complex formation. Furthermore, the quantity of hydrogen bonds (H-bond) was analyzed. As expected, a few numbers of averaged H-bond interactions were found in the vicinity of ~ 1–3 bonds, occurring ~ 10–47% during the entire simulated time. Following energy calculations (Table 3) and post-MD binding mode (Fig. 5), it becomes evident that the inhibitory action of all screened compounds was mainly driven by ensembles of hydrophobic interactions rather than electrostatic forces and H-bonds. In addition, we measured the solvent-accessible surface area (SASA) within the 5 Å sphere around the ligand. It was found that the SASA was in the range of 661–810 Å2 (662 Å2, 680 Å2, 810 Å2, and 756 Å2 for prazosin, rilpivirine, doxazosin, and nicergoline, respectively). The highest SASA value was observed in the case of doxazosine. Having higher polar surface areas may interrupt the hydrophobic/nonpolar stabilization50, resulting in reduced total binding affinity and the highest reaction fields (ΔGRF) observed (Table 3).
System stability and structural dynamics of the drug-protein complex. (A) root-mean-square deviation, RMSD; (B) time evolution of numbers of contact atoms; (C) solvent accessible surface area (SASA) measured the amino acids within the 5 Å around the compound candidates; (D) numbers of intermolecular hydrogen bonds (H-bonds) of all screened compounds, highlighting in different line colors.
Hot-spot residues for drug binding
To elucidate the amino-acid residues largely responsible for drug binding, the calculation of free energy decomposition (\(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\)) was performed using the MM/GBSA approach. We considered only residues showing \(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\) < -0.50 kcal/mol as “hot-spot” ones. There were 8–10 key binding amino acids found to be largely involved in compound recognition (Fig. 4). Specifically, some residues commonly interacted with almost all screened compounds including Leu380, Ser388, Leu394, Phe504, and Ile508, which align with regions observed in the inhibitors of the FZD7 transmembrane domain51. Additionally, residues such as Tyr385 and Ser388 significantly contributed to prazosin binding, while Tyr218 and Phe504 played crucial roles in rilpivirine binding. Notably, Met389 noticeably stabilized doxazosin recognition, exhibiting the highest \(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\) among all residues. In the case of nicergoline, Phe504 and Leu511 were important in the molecular recognition process. These identified residues are crucial hot-spot amino acids governing compound binding, warranting further exploration for the development of effective inhibitors targeting FZD10.
Hot-spot residues for drug binding to FZD10. (A) Free energy decomposition (\(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\)) of amino-acid residues largely contributed to the binding of candidate compounds; (B) Lists of key residues and their levels of contribution, showing the commonly and uniquely interacting residues among drug candidates. Note that only amino acid residues 100–581 were shown.
Post-MD binding mode analysis
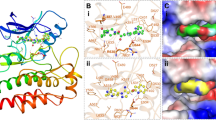
We employed the post-MD snapshot to observe the compounds’ binding mode and gain insights into protein-ligand recognition and susceptibility toward FZD10. Three main types of non-covalent interactions are responsible for stabilizing protein-ligand complexes, including van der Waals (vdW), conventional hydrogen bonds (H-bonds), and hydrophobic interactions (Pi-pi stacked, pi-pi T-shaped, Amide-pi stacked, pi-alkyl, and alkyl-alkyl) (Fig. 5). These interactions were visualized using the Discovery Studio 2017 program. The inhibitory actions primarily depended on hydrophobic interactions, which correlated well with the dominant van der Waals energy (EVdW) contributions to ΔGbind (Table 2). Additionally, the specific interactions of each compound were analyzed. Prazosin forms five hydrophobic interactions with Tyr385, Leu394, Val398, Lys507, and Ile508 (Fig. 5A), while rilpivirine hydrophobically interacts with Val215, Tyr218, Val391, Met469, Trp472, Lys473, Ala476, Pro499, and Phe504. However, some of them (Val215, Val391, Met469, and Ala476) might be weak and/or transient for binding since their \(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\) value was not noticeable (Fig. 5B). In the case of doxazosin, six amino acids form hydrophobic interactions including Met389, Trp472, Lys473, Ala476, Phe504, and Ile508. Interestingly, Met389 was considered a crucial residue owing to its highest \(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\) for doxazosin binding (Fig. 4A and 5C) and may be a reason advocating the high binding strength. Moreover, as expected, only a few H-bonds were observed in prazosin (TYR277 and SER388), rilpivirine (MET389), and doxazosin (Met389 and Asp390). Nicergoline shows high numbers of amino acids participating in the binding process, including Tyr218, Leu380, Tyr385, Val398, Met389, Phe504, Ile508, and Leu511 by hydrophobic interactions (Fig. 5D), which support the increased ΔGbind value.
Post-MD binding mode analysis of drug candidates. 2D interaction plots of compounds’ binding mode within the binding pocket of FZD10, utilizing the last MD snapshot as representative intermolecular interactions. The types of non-covalent interactions were shown in different line colors. (A) Prazosin; (B) Rilpivirine; (C) Doxazosin; (D) Nicergoline.
Assessment of the interaction between the repurposed drugs and FZD10 by STD-NMR
To experimentally validate the interaction between the selected repurposed drugs and FZD10, we employed saturation transfer difference nuclear magnetic resonance spectroscopy (STD-NMR). In this technique, resonances of the protein are saturated, and a ligand that binds to the protein will receive saturation to the protons near the protein. Once the ligand dissociates, a reduction in the NMR signal intensities can be observed. The difference between the signal intensities in the 1H NMR spectra with and without saturation is reported as an STD spectrum, which only gives the proton signals of dissociated ligands that have interacted with the protein. FZD10 and the respective drugs were diluted in D2O and subjected to STD-NMR analyses. The STD signals, corresponding to the proton signals of the drugs (Fig. 6) serve as indicators of the spatial proximity between these protons and FZD10, thereby signifying the binding of the drugs to FZD10.
STD-NMR validation of the interaction between drugs and FZD10. STD-NMR spectra of sample containing FZD10 recombinant proteoliposome with (A) Prazosin; (B) Rilpivirine; (C) Doxazosin; and (D) Nicergoline. Red: STD difference spectra acquired with 8 scans for an experimental time of 6 min 41 s. Black: 1D 1H NMR spectra acquired with a 30°pulse, without water suppression.
The STD-NMR findings partially support the non-covalent interactions postulated by post-MD binding mode predictions and the subsequent identification of key binding residues for drug interactions. In the case of nicergoline, the STD signals corresponding to H2 and H3 corroborate the hydrophobic interactions involving Ile508 (Figs. 5 and 6A). For doxazosin, the observed STD signals for H5 and H6 align with the hydrophobic interaction with the hotspot residue Met389 (Figs. 5 and 6B). The STD signals from H2 in rilpivirine supports its interaction with Phe504, a hotspot residue contributing significantly to \(\:{{\Delta\:}\text{G}}_{\text{r}\text{e}\text{s}\text{i}\text{d}\text{u}\text{e}}^{\text{b}\text{i}\text{n}\text{d}}\) (Figs. 4 and 5, and 6C). The STD signals from H2, H3,4, and H5,6 correspond to the predicted interactions between prazosin and Leu380, Ser388, and Leu394, respectively (Figs. 5 and 6D). It is noteworthy that STD signals also indicate potential interactions between the drugs and non-hotspot residues. For instance, in nicergoline, STD-signal from H4 potentially indicates hydrophobic interaction with Thr381 (Fig. 6A). Similarly, the STD signals for H3 and H4 in doxazosin might be attributed to interactions with Ile498 (Fig. 6B). Lastly, the STD signals for H1 and H5,6 in prazosin might signify potential interactions with Val391 and Asn378, respectively (Fig. 6D). The STD-NMR results largely confirmed our computational predictions on how the repurposed drugs interact with FZD10, validating the key binding modes and interactions, such as hydrophobic and hydrogen bonding with specific residues. The observed STD-NMR data complement MD analyses and provide additional insights into interactions that extend beyond MD prediction.
Effect of repurposed drugs on NPC cell growth
The study examined the expression of the FZD10 protein in normal nasopharyngeal and NPC cells. We observed the expression of FZD10 in both SUNE5-8F and SUNE6-10B NPC cells, however, the expression levels in NP460, normal nasopharyngeal cells, appeared to be extremely low (Fig. 7A). We further investigated the impact of drugs on NPC cell viability. The results showed the inhibitory effects of all tested drugs on both NPC cell lines (Fig. 7B). To strengthen our assessment of the cytotoxic effects observed in the MTT assays, we included the clonogenic assay, a well-established long-term viability assay. This method evaluates the capacity of single cells to survive, reproduce, and form colonies following drug treatment, offering a more comprehensive view of the repurposed drugs’ impact on NPC cell growth and proliferation over an extended period (Fig. 7C). Prazosin, at a concentration of 8 µM, inhibited colony formation in SUNE5-8F but not in SUNE6-10B. The IC50 dose of rilpivirine (4 µM) reduced the colony formation ability of both cell lines, while its IC25 dose (2 µM) had no impact on SUNE5-8F colony formation. In contrast, both IC25 and IC50 doses of doxazosin (7 and 14 µM) and nicergoline (20 and 40 µM) suppressed the colony formation of both cell lines. Our clonogenic assay results supported the findings from the MTT assay, providing robust confirmation of sustained cytotoxic effects of the four effective repurposed drugs on NPC cell lines. The consistent reduction in colony formation across treatment groups strongly suggests that these repurposed drugs exhibited a dose-dependent growth inhibition of NPC cells, and the observed cytotoxicity is not transient but likely results in long-term inhibition of cell viability and proliferation. It has been reported that the identified FDA-approved drugs herein exert effects on other cancer cells. For instance, prazosin has been shown to sensitize prostate cancer cells to chemotherapeutic drugs like docetaxel, suggesting its potential use in combination treatments for advanced prostate cancer44. Additionally, it inhibits the proliferation and viability of acute myeloid leukemia cells through the PI3K/Akt/mTOR signaling pathway45. Rilpivirine, a non-nucleoside reverse transcriptase inhibitor (NNRTI), has demonstrated toxicity to pancreatic cancer cells46. Doxazosin can induce apoptosis in various cancer cell types, including prostate cancer cells47, neuroblastoma cells48, breast cancer cell through EGFR and NF-κB pathway49. It also sensitizes cancer cells to Osimertinib, an anticancer drug50.
Effect of drug candidates on NPC cell growth. (A) The expression of FZD10 proteins in normal nasopharyngeal (NP460) and NPC (SUNE5-8F and SUNE6-10B) cells; (B) NPC cells were exposed to varying concentrations of drugs for 48 h, which revealed the cytotoxicity effect on both cells. (C) The clonogenic assay demonstrates the inhibitory effects of prazosin, rilpivirine, doxazosin, and nicergoline on the colony formation of SUNE5-8F and SUNE6-10B cells. (*p < 0.05 compared to the untreated control)
Effects of drugs on canonical Wnt signaling in NPC cells
Previous studies have demonstrated the role of FZD10 in transducing signals through the Wnt/β-catenin pathway and its involvement in NPC progression. The inhibitory effects of repurposed drugs on the FZD10 protein can be observed through the altered expression of the Wnt/β-catenin signaling pathway. The transduction of the canonical Wnt signaling pathway requires the interaction between the Fzd receptor and the corresponding Wnt ligand, which initiates a cascade of signaling events involving various proteins. This can be investigated through Western immunoblotting analysis. As anticipated, the expression of phosphorylated LRP6 and active β-catenin (non-phosphorylated β-catenin) proteins decreased after treatment with IC25 and IC50 doses of prazosin, rilpivirine, doxazosin, and nicergoline in SUNE5-8F cells (Fig. 8A). This suggests that these drugs may obstruct FZD10 from binding to its corresponding ligand, thereby reducing the transduction of canonical Wnt signaling in SUNE5-8F cells. In SUNE6-10B cells, the expression level of active β-catenin proteins decreased after treatment with the IC50 dose of prazosin, rilpivirine, and nicergoline. However, the expression of phosphorylated LRP6 was only reduced after exposure to the IC50 dose of rilpivirine. To specifically examine the expression of active β-catenin, nuclear non-phosphorylated β-catenin was quantified after treatment with the IC50 dose of these drugs through immunofluorescence. The results confirmed that all drug candidates significantly decreased the nuclear localization of non-phosphorylated β-catenin in both NPC cells (Fig. 8B). This experimental intensitometric analysis of phosphorylated LRP6 and non-phosphorylated β-catenin expression levels aligns with computational predictions, supporting the potential of these compounds to bind and inhibit FZD10 activity in NPC cell line models.
Effect of drug candidates on Wnt signaling proteins. (A) Expression of phosphorylated LRP6 and non-phosphorylated β-catenin in NPC cells after treatment with the IC25 and IC50 doses of prazosin, rilpivirine, doxazosin, and nicergoline for 48 h. Ten micrograms of each sample were loaded. The β-Actin serving as the loading control. The level of protein expression was quantified by intensitrometric analysis relative to actin as ratios to the untreated control. (B) Expression and localization of non-phosphorylated β-catenin in NPC cells using confocal fluorescence microscopy. Nuclear non-phosphorylated β-catenin (Non-P β-cat) was quantified by calculating corrected total cell fluorescence (CTCF) using ImageJ. (*p < 0.05, **p < 0.01 compared to untreated control), Scale bars, 20 μm.
Conclusion
This study presents a comprehensive approach to targeting FZD10 in NPC through drug repurposing. By constructing a reliable 3D model of FZD10 and utilizing molecular docking and dynamics simulations, we identified several FDA-approved drugs with potential inhibitory effects on FZD10. Among the drugs screened, nicergoline, doxazosin, rilpivirine, and prazosin showed significant binding affinity and selectivity towards FZD10, suggesting their potential for therapeutic application in NPC treatment. Molecular dynamics simulations revealed that these compounds predominantly interact with FZD10 through hydrophobic interactions, supported by van der Waals forces, rather than electrostatic or hydrogen bonding interactions. The stability of these complexes and the identification of key binding residues highlight the efficacy of these repurposed drugs in potentially disrupting the FZD10-mediated Wnt signaling pathway. Consistent with the cytotoxicity results, these four compounds were predicted to be well buried within the binding pocket of FZD10, potentially blocking its function and leading to reduced NPC cells growth. Detailed analysis of the binding modes and hot-spot residues further elucidated the mechanisms of inhibitory action, supported by STD-NMR experiments. The selected drugs also impacted the canonical Wnt signaling pathway, as evidenced by the reduction of phosphorylated LRP6 and non-phosphorylated β-catenin levels in NPC cells —a crucial pathway involved in NPC progression. This study underscores the promise of drug repurposing as a cost-effective strategy for developing targeted therapies against NPC, which remains a challenging cancer to treat due to its complex biological characteristics and therapeutic limitations. The insights gained from this research provide a foundation for further experimental validation and optimization of these drug candidates, paving the way for novel therapeutic approaches in cancer treatment. For example, the drug combination studies with the traditional chemotherapeutic agents, to validate these drugs as viable therapeutic options for NPC patients. The use of repurposed drugs for cancer treatment offers significant benefits, including reduced costs, lower side effects, and more efficient targeted therapy. However, long-term use of these drugs may lead to drug resistance over time. Consequently, repurposed drugs could serve as a viable option for cancer treatment, particularly when used in combination with traditional chemotherapeutic agents.
Data availability
Data will be made available on request to Tavan Janvilisri.
References
Liu, J. et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target. Therapy. 7 (1), 3 (2022).
Huang, X. et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J. Biol. Chem., (2018).
Li, G. et al. Frizzled7 Promotes Epithelial-to-mesenchymal Transition and Stemness Via Activating Canonical Wnt/beta-catenin Pathway in Gastric Cancer. Int. J. Biol. Sci. 14 (3), 280–293 (2018).
Stewart, D. J. Wnt signaling pathway in non-small cell lung cancer. J. Natl. Cancer Inst. 106 (1), djt356 (2014).
Zeng, Z. Y. et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum. Pathol. 38 (1), 120–133 (2007).
Staal, F. J. et al. Aberrant Wnt Signaling in Leukemia. Cancers (Basel), 8(9). (2016).
Peng, Q. et al. Overexpression of FZD1 is Associated with a Good Prognosis and Resistance of Sunitinib in Clear Cell Renal Cell Carcinoma. J. Cancer. 10 (5), 1237–1251 (2019).
Kirikoshi, H., Sekihara, H. & Katoh, M. Up-regulation of Frizzled-7 (FZD7) in human gastric cancer. Int. J. Oncol. 19 (1), 111–115 (2001).
Cao, T. T. et al. FZD7 is a novel prognostic marker and promotes tumor metastasis via WNT and EMT signaling pathways in esophageal squamous cell carcinoma. Oncotarget 8 (39), 65957–65968 (2017).
Yang, Q. et al. Frizzled 8 promotes the cell proliferation and metastasis of renal cell carcinoma. Oncotarget 8 (45), 78989–79002 (2017).
Koike, J. et al. Molecular cloning of Frizzled-10, a novel member of the Frizzled gene family. Biochem. Biophys. Res. Commun. 262 (1), 39–43 (1999).
Scavo, M. P. et al. Frizzled-10 and cancer progression: Is it a new prognostic marker? Oncotarget 9 (1), 824–830 (2018).
Tulalamba, W. et al. Transcriptomic profiling revealed FZD10 as a novel biomarker for nasopharyngeal carcinoma recurrence. Front. Oncol. 12, 1084713 (2022).
Gurney, A. et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl. Acad. Sci. U S A. 109 (29), 11717–11722 (2012).
Sudo, H. et al. FZD10-targeted α-radioimmunotherapy with (225) Ac-labeled OTSA101 achieves complete remission in a synovial sarcoma model. Cancer Sci. 113 (2), 721–732 (2022).
Chang, E. T. et al. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomarkers Prev. 30 (6), 1035–1047 (2021).
Paiar, F. et al. Role of chemotherapy in nasopharyngeal carcinoma. Oncol. Rev. 6 (1), e1 (2012).
Chan, A. T. et al. Overall survival after concurrent cisplatin-radiotherapy compared with radiotherapy alone in locoregionally advanced nasopharyngeal carcinoma. J. Natl. Cancer Inst. 97 (7), 536–539 (2005).
Nukulkit, S. et al. Maeruines A – E, elusive indole alkaloids from stems of Maerua siamensis and their inhibitory effects on cyclooxygenases and HT-29 colorectal cancer cell proliferation. Phytochemistry 229, 114291 (2025).
Shamsudin, N. F. et al. A novel chromone-based as a potential inhibitor of ULK1 that modulates autophagy and induces apoptosis in colon cancer. Future Medicinal Chemistry: pp. 1–19.
Leechaisit, R. et al. Novel Naphthoquinones as Potent Aromatase Inhibitors: Synthesis, Anticancer, and In silico studies. J. Mol. Struct., : p. 138981. (2024).
Suriya, U., Mahalapbutr, P. & Rungrotmongkol, T. Integration of In Silico Strategies for Drug Repositioning towards P38α Mitogen-Activated Protein Kinase (MAPK) at the Allosteric Site. Pharmaceutics 14 (7), 1461 (2022).
Suriya, U. et al. Quinoxalinones as a novel inhibitor scaffold for EGFR (L858R/T790M/C797S) tyrosine kinase: Molecular docking, biological evaluations, and computational insights. Molecules 27 (24), 8901 (2022).
Thirunavukkarasu, M. K. et al. In silico screening of available drugs targeting non-small cell lung cancer targets: A drug repurposing approach. Pharmaceutics 14 (1), 59 (2021).
Nalinratana, N. et al. In vitro and in silico studies of 7′′, 8′′-buddlenol D anti-inflammatory lignans from Carallia brachiata as p38 MAP kinase inhibitors. Sci. Rep. 13 (1), 3558 (2023).
Suriya, U. et al. Discovery of furopyridine-based compounds as novel inhibitors of Janus kinase 2: In silico and in vitro studies. Int. J. Biol. Macromol. 260, 129308 (2024).
Hengphasatporn, K. et al. Halogenated Baicalein as a Promising Antiviral Agent toward SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 62 (6), 1498–1509 (2022).
Nutho, B. et al. Discovery of C-12 dithiocarbamate andrographolide analogues as inhibitors of SARS-CoV-2 main protease: In vitro and in silico studies. Comput. Struct. Biotechnol. J. 20, 2784–2797 (2022).
Pojtanadithee, P. et al. Identification of Promising Sulfonamide Chalcones as Inhibitors of SARS-CoV-2 3CLpro through Structure-Based Virtual Screening and Experimental Approaches. J. Chem. Inf. Model. 63 (16), 5244–5258 (2023).
Suriya, U. et al. A diarylheptanoid derivative mediates glycogen synthase kinase 3β to promote the porcine muscle satellite cell proliferation: Implications for cultured meat production. Biochem. Biophys. Res. Commun. 736, 150850 (2024).
Bhardwaj, V. et al. Elimination of bitter-off taste of stevioside through structure modification and computational interventions. J. Theor. Biol. 486, 110094 (2020).
Purohit, R. et al. Studies on flexibility and binding affinity of Asp25 of HIV-1 protease mutants. Int. J. Biol. Macromol. 42 (4), 386–391 (2008).
Tanwar, G. & Purohit, R. Gain of native conformation of Aurora A S155R mutant by small molecules. J. Cell. Biochem. 120 (7), 11104–11114 (2019).
K, B. & Purohit, R. Mutational analysis of TYR gene and its structural consequences in OCA1A. Gene 513 (1), 184–195 (2013).
Kumar, A. & Purohit, R. Computational investigation of pathogenic nsSNPs in CEP63 protein. Gene 503 (1), 75–82 (2012).
Ashburn, T. T. & Thor, K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3 (8), 673–683 (2004).
Kelley, L. A. et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 (6), 845–858 (2015).
Wallner, B. & Elofsson, A. Can correct protein models be identified? Protein Sci. 12 (5), 1073–1086 (2003).
Schneidman-Duhovny, D. et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res., (2005). 33(Web Server issue): pp. W363–W367 .
Yang, J. M. & Chen, C. C. GEMDOCK: a generic evolutionary method for molecular docking. Proteins 55 (2), 288–304 (2004).
Kramer, B., Rarey, M. & Lengauer, T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins 37 (2), 228–241 (1999).
Guzman, C. et al. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 9 (3), e92444 (2014).
Wang, J., Wang, J. & Caldwell, J. W., Romain M. Wolf, James W. Caldwell, Peter A. Kollman, and David A. Case, Development and testing of a general amber force field Journal of Computational Chemistry (2004) 25 (9) 1157–1174. 26(1): pp. 114–114. (2005).
Jorgensen, W. L. et al. Comparison simple potential. Funct. simulating liquid water 79(2): 926–935. (1983).
Darden, T. & York, D. and L.J.T.J.o.c.p. Pedersen, Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. 98(12): pp. 10089–10092. (1993).
Ryckaert, J. P. & Ciccotti, G. J.J.J.o.c.p. Berendsen. Numer. Integr. cartesian equations motion Syst. constraints: Mol. dynamics n-alkanes. 23 (3), 327–341 (1977).
Uberuaga, B. P. & Anghel, M. and A.F.J.T.J.o.c.p. Voter, Synchronization of trajectories in canonical molecular-dynamics simulations: Observation, explanation, and exploitation. 120(14): pp. 6363–6374. (2004).
Berendsen, H. J. et al. Molecular dynamics with coupling to an external bath. 81(8): pp. 3684–3690. (1984).
Lica, J. J. et al. Effective Drug Concentration and Selectivity Depends on Fraction of Primitive Cells. Int. J. Mol. Sci., 22(9). (2021).
Wang, J. et al. Solvation Model Based on Weighted Solvent Accessible Surface Area. J. Phys. Chem. B. 105 (21), 5055–5067 (2001).
Zhang, W. et al. Discovery of novel frizzled-7 inhibitors by targeting the receptor’s transmembrane domain. Oncotarget; Vol 8, No 53, (2017).
Acknowledgements
The authors express their gratitude to the Research Unit for Natural Product Biotechnology, Faculty of Pharmaceutical Sciences, Chulalongkorn University, and to Prof. Wanchai De-Eknamkul for providing the FlexX software and technical assistance with LeadIT. We also thank the Central Instrument Facility at Faculty of Science, Mahidol University. T.R. thanks Chulalongkorn University (CE66_036_2300_008) for support.
Funding
This research project is supported by Mahidol University (Fundamental Fund: fiscal year 2023 by National Science Research and Innovation Fund (NSRF)).
Author information
Authors and Affiliations
Contributions
CN: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. US: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. PP: Formal analysis, Investigation. KV: Investigation. TR: Methodology, Validation, Resources, Writing - Review & Editing. TS: Methodology, Investigation. SK: Methodology, Validation, Formal analysis, Investigation, Writing - Review & Editing. TJ: Conceptualization, Validation, Resources, Writing - Review & Editing, Supervision, Project administration, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ngernsombat, C., Suriya, U., Prattapong, P. et al. Repurposing FDA-approved drugs targeting FZD10 in nasopharyngeal carcinoma: insights from molecular dynamics simulations and experimental validation. Sci Rep 14, 31461 (2024). https://doi.org/10.1038/s41598-024-82967-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82967-7